Histone Ubiquitylation
- Search List
- Background
- Molecules
- Mechanism
- Disease
- Case Study
Related Symbol Search List
Immunology Background
Background
Histone ubiquitylation is a post-translational modification process that involves the addition of ubiquitin molecules to histone proteins. Ubiquitin is a small protein that can be attached to target proteins, thereby affecting their stability, localization, and function. Histone ubiquitylation plays a crucial role in regulating chromatin structure, gene expression, DNA repair, and other important cellular processes.
The Key Functions and Effects of Histone Ubiquitylation
Functionally, histone ubiquitylation can have diverse effects on chromatin and gene regulation. Here are some key functions and effects of histone ubiquitylation:
Transcriptional regulation
Histone ubiquitylation can modulate gene expression by influencing the accessibility of DNA to transcription factors and the transcription machinery. It can act as both a positive and negative regulator of transcription, depending on the specific histone residue and the associated proteins involved.
DNA damage response
Histone ubiquitylation is involved in the response to DNA damage. It helps recruit DNA repair factors to the damaged sites, facilitating the repair process. It also plays a role in signaling and coordinating DNA damage checkpoints and cell cycle arrest.
Chromatin remodeling
Histone ubiquitylation can affect higher-order chromatin structure and dynamics. It can regulate the activity of chromatin remodeling complexes, such as SWI/SNF, by promoting their recruitment or activation at specific genomic loci.
Epigenetic inheritance
Histone ubiquitylation can contribute to the establishment and maintenance of epigenetic states. Certain histone ubiquitylation marks serve as docking sites for other effector proteins that help propagate specific chromatin states through cell divisions.
Protein-protein interactions
Histone ubiquitylation can promote the recruitment of various proteins involved in chromatin regulation and DNA repair. It acts as a binding platform for effector proteins containing ubiquitin-binding domains, facilitating their association with specific genomic regions.
Crosstalk with other histone modifications
Histone ubiquitylation can interact with and influence the effects of other histone modifications, such as acetylation, methylation, and phosphorylation. This crosstalk between different histone modifications adds another layer of complexity to chromatin regulation and gene expression.
The Role of Histone Ubiquitylation in Various Aspects
Histone ubiquitylation plays a critical role in cell cycle regulation, DNA repair, and chromosome structure maintenance. Here's an overview of its involvement in these processes:
Cell cycle regulation
Histone ubiquitylation is intricately linked to cell cycle progression and the coordination of different cell cycle phases. Ubiquitylation of histones regulates the accessibility of DNA and the recruitment of various proteins involved in cell cycle control. For example:
- H2BK120ub1: This modification is associated with active transcription elongation and is important for proper cell cycle progression. It facilitates efficient mRNA processing and is involved in the expression of genes that regulate cell cycle checkpoints.
- SCF complexes: These E3 ubiquitin ligase complexes target cell cycle regulators, such as cyclins and cyclin-dependent kinase inhibitors (CDKIs), for ubiquitylation and subsequent degradation. This process ensures the timely transition between different cell cycle phases.
DNA repair
Histone ubiquitylation plays a crucial role in DNA repair processes by regulating the recruitment of repair factors and the assembly of repair complexes at sites of DNA damage. Some key examples include:
- H2BK123ub1: This modification is associated with DNA double-strand break repair. It facilitates the recruitment of repair factors, such as the BRCA1 and 53BP1 proteins, to the damaged sites, promoting efficient repair.
- H2AK13/15ub1: This modification is involved in the recognition and repair of DNA lesions. It recruits DNA repair factors, including the nucleotide excision repair (NER) machinery, to sites of DNA damage.
Chromosome structure maintenance
Histone ubiquitylation is essential for the maintenance of chromatin structure and organization, which is critical for proper chromosome function. It helps establish distinct chromatin domains and regulates higher-order chromatin structures. Notable examples include:
- H2AK119ub1: This modification is catalyzed by the Polycomb Repressive Complex 1 (PRC1) and contributes to gene silencing and the maintenance of repressive chromatin states. It plays a role in maintaining chromatin structure and preventing the spread of active chromatin marks.
- H2BK120ub1: Besides its role in transcriptional regulation, this modification also participates in maintaining proper chromosome structure. It influences chromatin compaction and organization, ensuring the stability and integrity of chromosomes.
Histone ubiquitylation is a dynamic and tightly regulated process that impacts various aspects of cell cycle regulation, DNA repair, and chromosome structure maintenance. Dysregulation of histone ubiquitylation can lead to genomic instability, defects in DNA repair, and aberrant chromatin organization, which are associated with various diseases, including cancer. Understanding the mechanisms and functional consequences of histone ubiquitylation in these processes contributes to our knowledge of fundamental cellular functions and has implications for therapeutic interventions targeting these pathways.
How Does Histone Ubiquitylation Affect Gene Expression and Transcriptional Regulation?
Histone ubiquitylation plays a crucial role in gene expression and transcriptional regulation by influencing chromatin structure, recruitment of transcriptional regulators, and the assembly of protein complexes. Here are some ways in which histone ubiquitylation affects gene expression:
- Chromatin remodeling: Histone ubiquitylation can alter the structure of chromatin, making it more or less accessible to transcriptional machinery. For example, monoubiquitylation of histone H2B at lysine 120 (H2BK120ub1) is associated with transcriptional activation and can lead to chromatin decompaction, allowing transcription factors and RNA polymerase II to access the DNA and initiate transcription.
- Recruitment of transcriptional regulators: Histone ubiquitylation can serve as a binding site for specific proteins involved in transcriptional regulation. For example, H2BK120ub1 recruits the E3 ligase RNF20, which in turn recruits the E3 ligase complex PRC1. These complexes can modulate chromatin structure and gene expression through interactions with other regulatory proteins.
- Crosstalk with other histone modifications: Histone ubiquitylation can crosstalk with other histone modifications, such as methylation, acetylation, and phosphorylation. This interplay between different histone modifications can influence the recruitment of chromatin remodeling complexes and transcriptional regulators, leading to either gene activation or repression.
- Regulation of transcriptional elongation: Histone ubiquitylation can impact transcriptional elongation, the process by which RNA polymerase II moves along the DNA template to synthesize RNA. H2BK120ub1, in particular, is associated with increased transcriptional elongation rates and efficient mRNA processing.
- Assembly of protein complexes: Histone ubiquitylation can facilitate the assembly of protein complexes involved in transcriptional regulation. For example, H2AK119ub1, catalyzed by the PRC1 complex, promotes the recruitment of other PRC1 subunits and PRC2 complex, leading to the establishment of repressive chromatin states and gene silencing.
- DNA repair and replication: Histone ubiquitylation is also involved in DNA repair and replication processes. It can regulate the recruitment of DNA repair factors and facilitate the progression of DNA replication forks.
Overall, histone ubiquitylation acts as a dynamic modification that regulates gene expression by modulating chromatin structure, recruiting transcriptional regulators, and influencing the assembly of protein complexes involved in transcriptional regulation. The specific outcomes of histone ubiquitylation depend on the context, the targeted histone residues, and the associated protein complexes.
Which Enzymes are Involved in Histone Ubiquitylation?
The regulatory mechanism of histone ubiquitylation involves a complex interplay between different enzymes that mediate the addition and removal of ubiquitin from histone proteins. The key enzymes involved in histone ubiquitylation are E3 ubiquitin ligases, deubiquitinases (DUBs), and ubiquitin-conjugating enzymes (E2).
| Enzymes | Details |
|---|---|
| E3 Ubiquitin Ligases |
E3 ligases play a central role in catalyzing the transfer of ubiquitin molecules to histone proteins. They recognize specific histone residues and facilitate the transfer of ubiquitin from E2 enzymes to the target histones. Different E3 ligases are associated with distinct histone residues and have specific functions. Some well-known E3 ligases involved in histone ubiquitylation include:
|
| Deubiquitinases (DUBs) |
DUBs are enzymes that remove ubiquitin from target proteins, including ubiquitylated histones. They counterbalance the ubiquitylation process and regulate the dynamic turnover of ubiquitin marks on histones. Some DUBs known to be involved in histone deubiquitylation include:
|
| Ubiquitin-Conjugating Enzymes (E2) |
E2 enzymes work in conjunction with E3 ligases to transfer ubiquitin molecules to target proteins. They interact with E3 ligases and facilitate the transfer of ubiquitin from E1 activating enzymes to the target histones. Different E2 enzymes are associated with specific E3 ligases and histone modifications.
|
These enzymes, along with other regulatory factors and cofactors, form a complex regulatory network that controls the precise patterns of histone ubiquitylation in response to various cellular signals and processes. The interplay between E3 ligases, DUBs, and E2 enzymes ensures the dynamic and reversible nature of histone ubiquitylation, allowing for fine-tuned chromatin regulation and gene expression control.
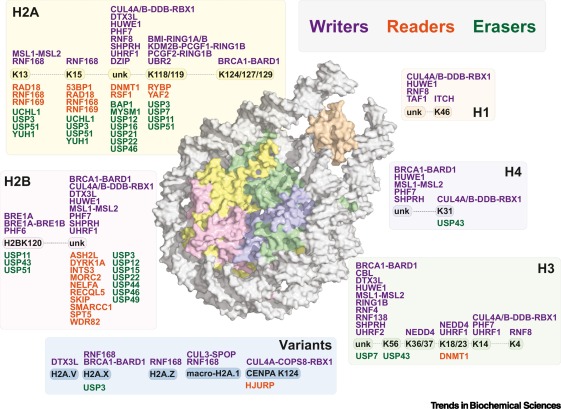 Fig.1 Regulators of histone ubiquitination. (Vaughan RM, et al., 2021)
Fig.1 Regulators of histone ubiquitination. (Vaughan RM, et al., 2021)Histone Ubiquitylation-Related Molecules
Histone ubiquitination is involved in the activity of various molecules involved in the ubiquitin-proteasome system and transcriptional regulation. The following is an introduction to some of the molecules associated with histone ubiquitination:
| Types | Details |
|---|---|
| E3 Ubiquitin Ligases |
|
| E2 Ubiquitin-Conjugating Enzymes |
|
| E1 Ubiquitin-Activating Enzymes |
|
| Ubiquitin-Activating Enzyme E1-like Protein |
|
| Transcriptional Regulators |
|
| Other Proteins |
|
These molecules collectively contribute to the dynamic regulation of histone ubiquitylation, which plays crucial roles in chromatin structure, gene expression, and various cellular processes.
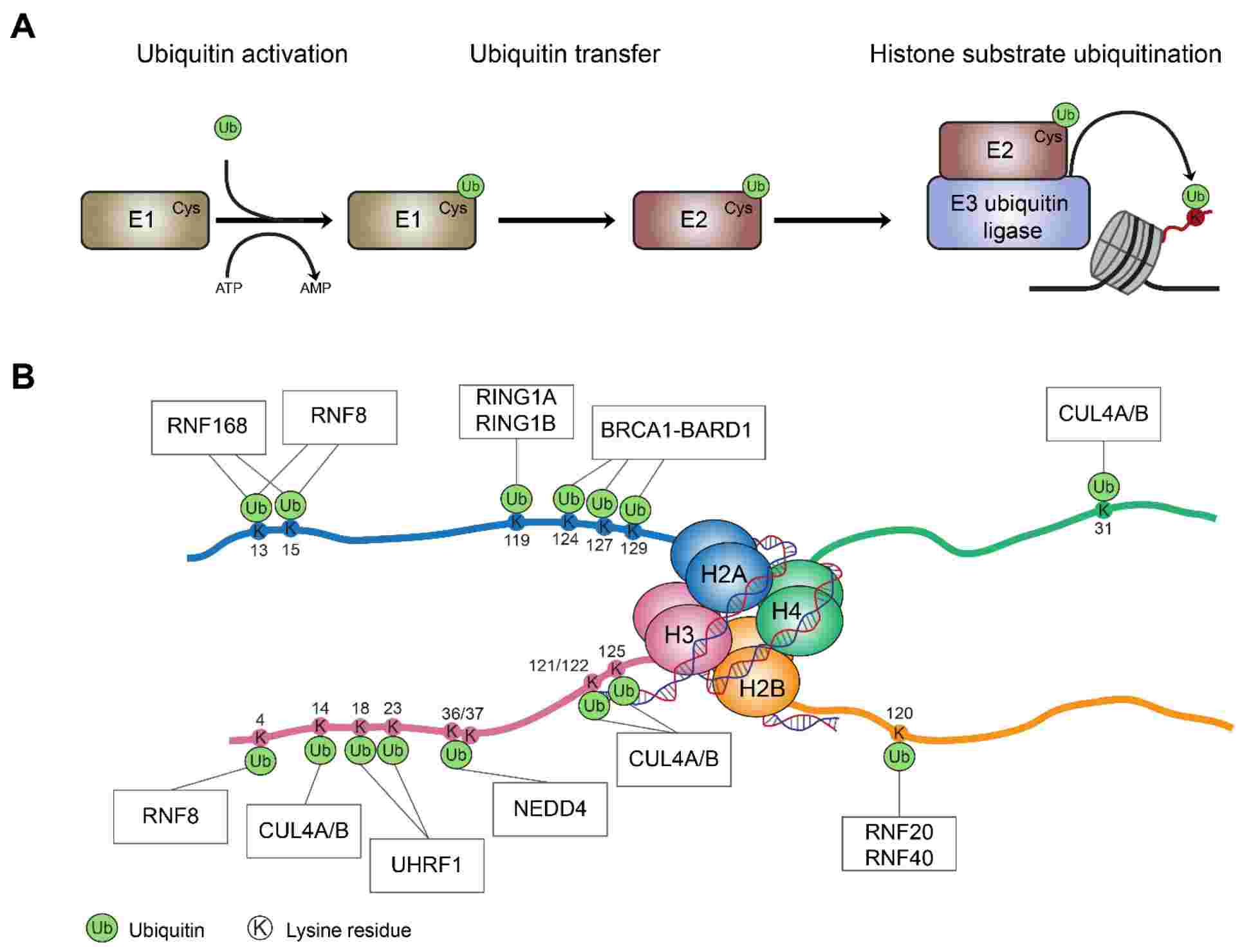 Fig.2 E3 ubiquitin ligases and histone ubiquitination events. (Oss-Ronen L, et al., 2022)
Fig.2 E3 ubiquitin ligases and histone ubiquitination events. (Oss-Ronen L, et al., 2022)The Regulatory Mechanism of Histone Ubiquitylation
The regulatory mechanism of histone ubiquitylation involves a complex interplay of various factors, including chromatin modifiers, transcriptional regulators, signaling pathways, and DNA damage responses. Here are some key regulatory mechanisms involved in histone ubiquitylation:
| Key regulatory mechanisms | Details |
|---|---|
| Chromatin context | The local chromatin environment and accessibility play a crucial role in determining the sites and extent of histone ubiquitylation. Chromatin modifiers, such as ATP-dependent chromatin remodeling complexes and histone acetyltransferases (HATs), can create an open chromatin state that facilitates the recruitment of E3 ubiquitin ligases to specific genomic regions. |
| Transcriptional regulation | Transcriptional activators and repressors can influence histone ubiquitylation by recruiting or modulating the activity of E3 ligases. For example, transcriptional activators may recruit E3 ligases to promote histone ubiquitylation, leading to gene activation. Conversely, repressors can inhibit or remove ubiquitin marks from histones, resulting in gene repression. |
| Signaling pathways | Various signaling pathways, such as those activated by growth factors, stress stimuli, or DNA damage, can regulate histone ubiquitylation. These pathways often involve protein kinases that phosphorylate histones or downstream targets, leading to changes in chromatin structure and the recruitment of E3 ligases or DUBs. |
| DNA damage response | DNA damage triggers a cascade of events that involves histone ubiquitylation. E3 ligases, such as RNF8 and RNF168, are recruited to DNA damage sites where they facilitate histone ubiquitylation, primarily on H2A and H2AX. This ubiquitylation serves as a recruitment platform for DNA repair factors and signaling proteins involved in the DNA damage response. |
| Crosstalk with other histone modifications | Histone ubiquitylation can interact with and be influenced by other histone modifications, including acetylation, methylation, and phosphorylation. Certain modifications can create binding sites for E3 ligases or alter the accessibility of histones to these ligases, thereby influencing the ubiquitylation process. |
| Dynamic turnover | Histone deubiquitinases (DUBs) play a critical role in regulating the dynamic turnover of ubiquitin marks on histones. DUBs remove ubiquitin from histones, counterbalancing the ubiquitylation process and allowing for precise control and reversible regulation of chromatin states. |
The regulatory mechanisms of histone ubiquitylation are highly complex and interconnected. They involve a combination of chromatin modifiers, transcriptional regulators, signaling pathways, and DNA damage responses. These mechanisms ensure the precise spatiotemporal control of histone ubiquitylation, allowing for dynamic chromatin remodeling and the regulation of various cellular processes, including gene expression, DNA repair, and epigenetic inheritance.
The Role of Histone Ubiquitylation in Disease Development and Potential Clinical Applications
Histone ubiquitylation plays important roles in various biological processes, and dysregulation of this modification has been implicated in the development of several diseases. Understanding the role of histone ubiquitylation in disease pathogenesis can potentially lead to the development of novel therapeutic strategies. Here are some examples of the involvement of histone ubiquitylation in disease development and potential clinical applications:
Cancer
Alterations in histone ubiquitylation have been observed in various types of cancer. Dysregulation of E3 ligases, such as BMI1 and RING1, can lead to aberrant histone ubiquitylation patterns, affecting gene expression and promoting oncogenesis. Targeting histone ubiquitylation machinery components or their downstream effectors provides potential avenues for cancer therapy.
Neurodegenerative diseases
Abnormal histone ubiquitylation has been implicated in neurodegenerative disorders such as Alzheimer's disease and Parkinson's disease. Dysregulation of E3 ligases and deubiquitinases involved in histone ubiquitylation can impact the expression of genes associated with neuronal function and survival. Modulating histone ubiquitylation may hold therapeutic potential for neurodegenerative diseases.
Epigenetic disorders
Histone ubiquitylation defects can contribute to epigenetic disorders. For example, mutations in the RNF20 gene, encoding an E3 ligase responsible for H2BK120ub1, have been associated with intellectual disability and developmental delay. Targeting histone ubiquitylation pathways could potentially correct epigenetic abnormalities and mitigate the symptoms of such disorders.
Therapeutic targeting
Understanding the molecular mechanisms of histone ubiquitylation has led to the development of potential therapeutic strategies. Small molecules or inhibitors targeting specific E3 ligases, deubiquitinases, or downstream effectors involved in histone ubiquitylation could be explored as therapeutic interventions for diseases associated with dysregulated histone modifications.
Biomarker discovery
Histone modifications, including ubiquitylation, have the potential to serve as biomarkers for disease diagnosis, prognosis, and treatment response. Analysis of histone ubiquitylation patterns could provide valuable insights into disease mechanisms and aid in the development of personalized treatment approaches.
It is important to note that the field of histone ubiquitylation and its clinical applications is still evolving, and further research is needed to fully understand its complexities and therapeutic potential. However, the study of histone ubiquitylation in disease contexts holds promise for advancing our understanding of disease mechanisms and developing innovative therapeutic interventions.
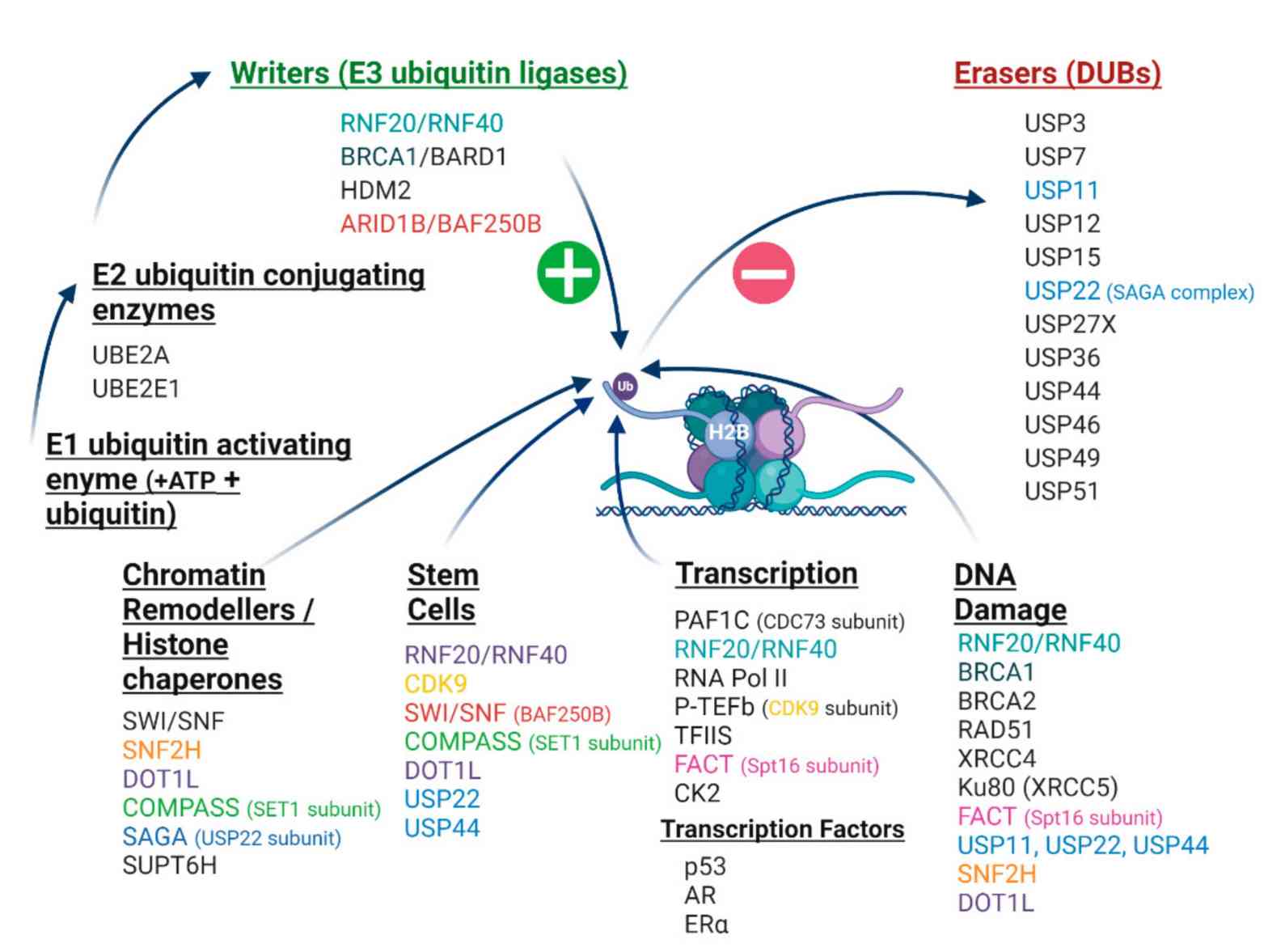 Fig.3 The H2Bub1 interactome. (Marsh DJ, et al., 2020)
Fig.3 The H2Bub1 interactome. (Marsh DJ, et al., 2020)Case Study
Case 1: Wang P, Guo K, Su Q, Deng J, Zhang X, Tu L. Histone ubiquitination controls organ size in cotton (Gossypium hirsutum). Plant J. 2022;110(4):1005-1020.
This study investigated the role of protein ubiquitination in organ size control in cotton (Gossypium hirsutum). The researchers identified an E2 enzyme called GhUBC2L and demonstrated its positive role in cell proliferation and expansion. Complete knock-down of GhUBC2L led to reduced growth and organ size, while overexpression of GhUBC2L promoted cotton growth and resulted in enlarged organs. Histone monoubiquitination of H2A and H2B was significantly impaired in GhUBC2L-suppressed cotton but slightly enhanced in GhUBC2L-overexpressing plants. The researchers also discovered that GhUbox8, a U-box type E3 ligase protein, interacted with GhUBC2L, suggesting their collaborative function in protein ubiquitination. Furthermore, GhUbox8 was found to interact with histone proteins, including H2A and H2B, indicating its potential role in monoubiquitination of these histones. The expression of genes related to cell cycle and organ development was altered when the expression of GhUBC2L was manipulated. These findings provide new insights into the regulation of organ size control through protein ubiquitination, particularly histone monoubiquitination, in plant development, specifically in cotton.
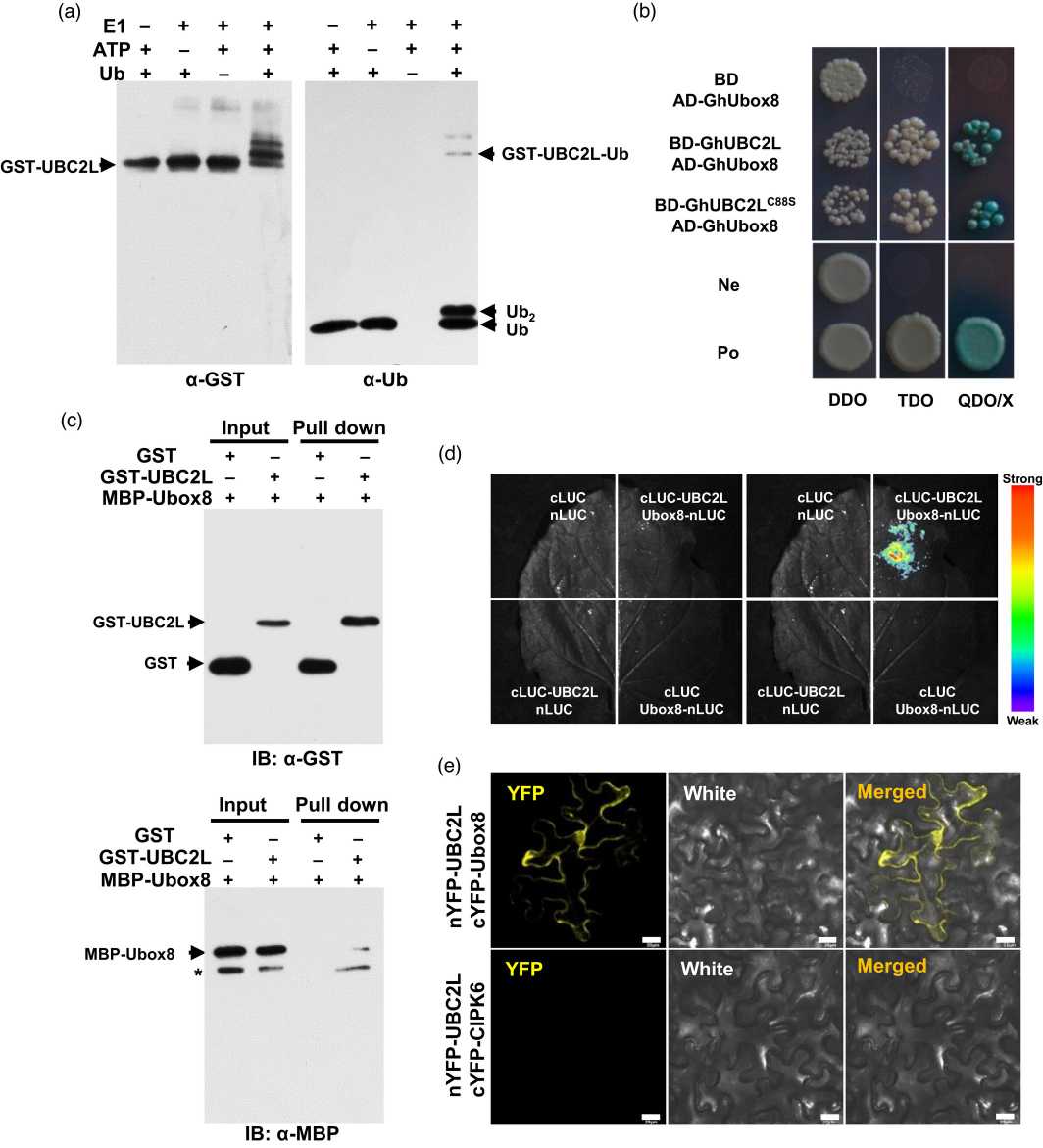 Fig.1 GhUBC2L is an active E2 and interacts with a U-box type E3 ligase GhUbox8.
Fig.1 GhUBC2L is an active E2 and interacts with a U-box type E3 ligase GhUbox8.Case 2: Wegwitz, F., Prokakis, E., Pejkovska, A. et al. The histone H2B ubiquitin ligase RNF40 is required for HER2-driven mammary tumorigenesis. Cell Death Dis 11, 873 (2020).
This study focused on the aggressive HER2-positive breast cancer subtype (HER2+-BC) and the development of new treatment strategies for patients who become refractory to current targeted therapies. The researchers investigated the role of the epigenetic regulator RNF40 and its association with histone 2B monoubiquitination (H2Bub1) in HER2+-BC. Previous studies suggested that loss of H2Bub1 is associated with cancer progression, but this study unexpectedly found a tumor-supportive role of RNF40 in HER2+-BC. Using human tumor samples, cell culture models, and a mouse model with Rnf40 deletion in mammary carcinoma tissue, the researchers demonstrated that RNF40-driven H2B monoubiquitination is essential for the transcriptional activation of components in the RHO/ROCK/LIMK pathway and proper actin-cytoskeleton dynamics. They also discovered a trans-histone crosstalk between H2B monoubiquitination and histone 3 lysine 4 trimethylation (H3K4me3). The findings suggest that the H2B monoubiquitination axis, regulated by RNF40, could be a tumor context-dependent therapeutic target in HER2+-BC, providing potential opportunities for developing new treatment approaches in breast cancer.
The test aimed to investigate the molecular mechanisms underlying the dependence of HER2-positive breast cancer (HER2+-BC) on RNF40. Two HER2+-BC cell lines, HCC1954 and SKBR3, were selected for the study. The researchers used siRNA-mediated knockdown of RNF40 to assess the impact on various tumorigenic properties of the cell lines. Depletion of RNF40 and loss of H2B monoubiquitination (H2Bub1) resulted in reduced cellular confluency, impaired growth kinetics, decreased clonogenic capacity, and inhibited tumorsphere formation in both cell lines. Analysis of gene essentiality data from the DepMap portal confirmed the strong dependence of HER2+-BC cell lines on RNF40 expression. Additionally, the levels of the proliferation marker Ki67 were significantly reduced upon RNF40 depletion. The migration potential of HCC1954 cells was also impaired upon RNF40 loss, as demonstrated by transwell migration and gap closure assays. These findings, combined with in vivo observations, indicate that RNF40 expression is essential for maintaining the tumorigenic properties of HER2+-BC cells both in vitro and in vivo.
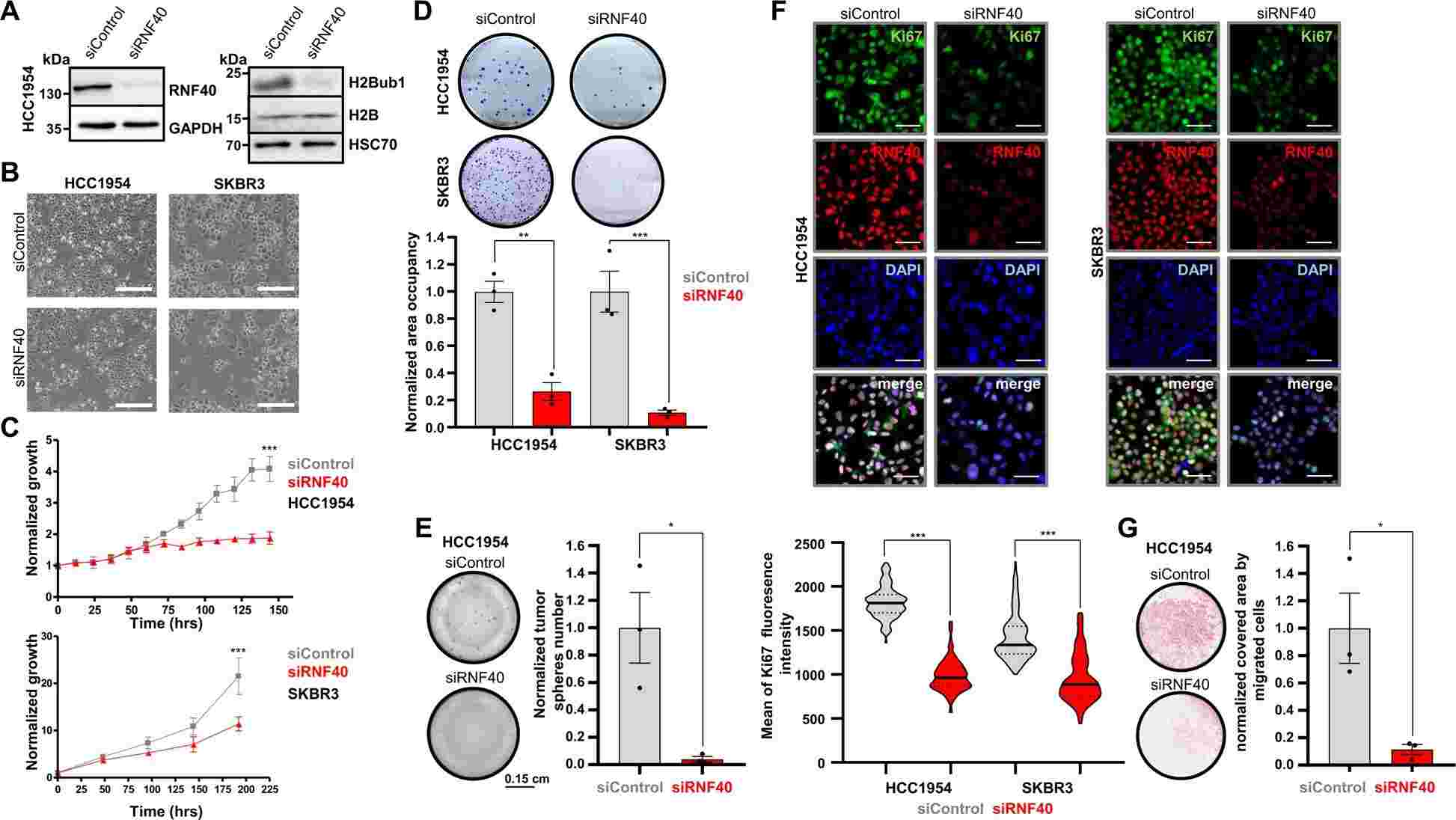 Fig.2 RNF40 loss impairs oncogenic properties of HER2-BC cells in vitro.
Fig.2 RNF40 loss impairs oncogenic properties of HER2-BC cells in vitro.Related References
- Chen JJ, Stermer D, Tanny JC. Decoding histone ubiquitylation. Front Cell Dev Biol. 2022;10:968398. Published 2022 Aug 29.
- Cao J, Yan Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front Oncol. 2012;2:26.
- Vaughan RM, Kupai A, Rothbart SB. Chromatin regulation through ubiquitin and ubiquitin-like histone modifications. Trends Biochem Sci. 2021;46(4):258-269.
- Oss-Ronen L, Sarusi T, Cohen I. Histone mono-ubiquitination in transcriptional regulation and its mark on life: emerging roles in tissue development and disease. Cells. 2022; 11(15):2404.
- Marsh DJ, Ma Y, Dickson K-A. Histone monoubiquitination in chromatin remodeling: focus on the histone H2B interactome and cancer. Cancers. 2020; 12(11):3462.
