Epigenetics-Based Assay Development
Background
Epigenetics, the study of heritable changes in gene expression that do not involve alterations to the underlying DNA sequence, has emerged as a crucial field in modern biology. It encompasses a range of molecular mechanisms, including DNA methylation, histone modification, and non-coding RNA regulation, which play vital roles in cellular differentiation, development, and disease pathogenesis. As our understanding of epigenetic processes deepens, the potential for leveraging these insights to develop innovative diagnostic and therapeutic tools becomes increasingly evident. Creative BioMart is at the forefront of this scientific revolution, committed to advancing the field of epigenetics through cutting-edge assay development.
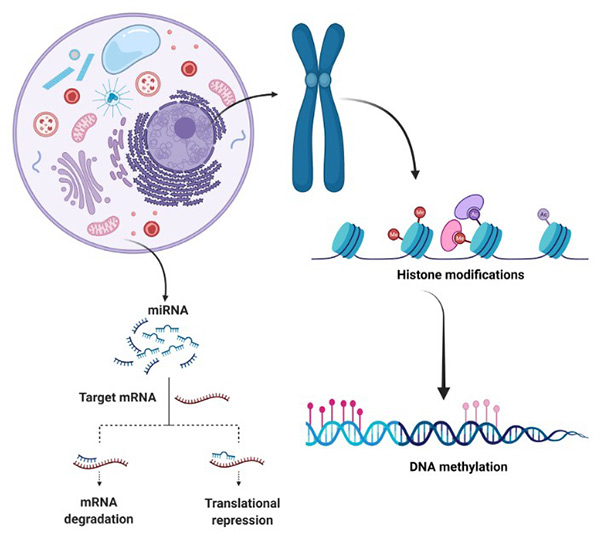
Figure 1. Main epigenetic mechanisms. (Borges, 2022)
What We Offer?
Creative BioMart offers comprehensive epigenetic solutions and services for assay development:
|
Type |
 Biochemical Assay Development |
 Biochemical Protein-Protein Interaction Assay Development |
 Cellular Assay Development |
|
Details |
Focus on enzyme activity with epigenetic targets (methyltransferases, demethylases, acetyltransferases, etc.) |
Focus on protein interactions in epigenetic pathways (reader domains, chromatin complexes) |
Focus on cellular phenotypes and epigenetic marker changes |
|
Assay formats |
TR-FRET (LANCE Ultra, HTRF), Caliper, fluorometric, luminescent, radiometric, etc. |
TR-FRET (HTRF, LANCE), fluorescence polarization, Biacore, radiometric, etc. |
BacMam, ELISA, Western blot, high-content imaging, flow cytometry, etc. |
|
Substrate choices |
Peptide, recombinant histone protein, reconstituted nucleosome, DNA/RNA oligonucleotides |
Recombinant epigenetic proteins, modified histone peptides, nucleosome complexes |
Cell lines (primary, immortalized), reporter cell lines |
|
Assay development |
Routine optimization (enzyme titration, time course, Km determination) and validation (DMSO tolerance, Z' test, compound validation) |
Routine optimization (protein titration, Kd determination, antibody titration) and validation |
Antibody specificity evaluation, routine optimization and validation, phenotypic screening |
|
Epigenetic Applications |
Histone modification assays, DNA methylation/demethylation assays, nucleosome remodeling |
Protein-protein interactions in chromatin regulation, epigenetic reader assays |
Cellular epigenetic marker detection, chromatin accessibility assays |
Why Choose Us?
- Expertise and Experience: Our skilled team brings deep knowledge in epigenetics, backed by a strong track record of developing innovative, application-specific assays.
- Cutting-Edge Technology: Equipped with the latest tools, our advanced labs ensure high-quality, reliable results using the most current epigenetic technologies.
- Customized Solutions: We tailor each assay to your unique research goals, offering flexible, client-focused development for optimal performance and efficiency.
- Quality and Reliability: All assays are rigorously validated for accuracy and reproducibility, ensuring dependable results for critical research decisions.
- Comprehensive Support: From design to data interpretation, our team offers full-service support, delivering responsive guidance at every stage of your project.
- Broad Assay Portfolio: Expertise across a wide range of epigenetic modifications—including DNA methylation, histone modifications, and chromatin accessibility.
Case Study
* NOTE: We prioritize confidentiality to safeguard our clients’ technology and intellectual property. As an alternative, we present selected published research articles as representative case studies. For details on the assay services and products used in these studies, please refer to the relevant sections of the cited literature.
Case 1: Enhancing chemotherapy response in AML via EZH2 inhibition and chromatin decondensation
Porazzi et al., 2022. doi:10.1158/0008-5472.CAN-21-1297
Acute myelogenous leukemia (AML) remains difficult to treat despite intensive chemotherapy. Researchers found AML cells rapidly accumulate the repressive histone mark H3K27me3 on nascent DNA. Inhibiting EZH2, the enzyme that catalyzes this mark, decondenses chromatin, increasing DNA accessibility and enhancing chemotherapy-induced DNA damage, apoptosis, and leukemia suppression. These effects were amplified when cells entered S-phase after CDK4/6-mediated G1 arrest. This strategy may improve treatment efficacy and allow lower chemotherapy doses, potentially reducing side effects, especially in older or vulnerable patients.
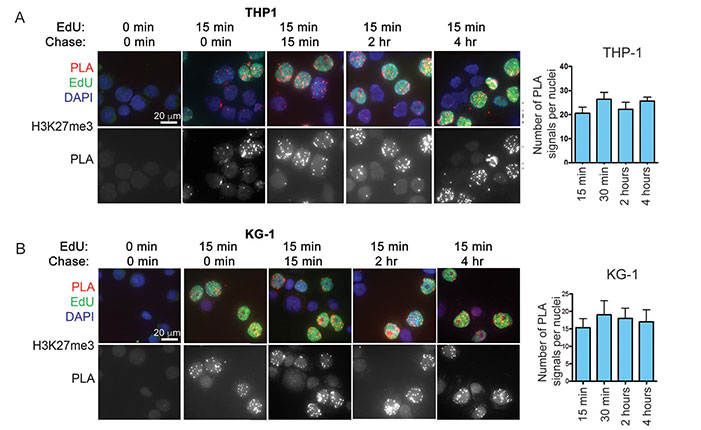
Figure 2. Accumulation of H3K27me3 on nascent DNA of AML cells. DNA of AML cell lines. (Porazzi et al., 2022)
Case 2: mqMSP assay enhances early detection and recurrence monitoring in colorectal cancer
Jin et al., 2020. doi:10.1073/pnas.2017421118
A novel multiplex methylation-specific qPCR assay (mqMSP) using 10 ctDNA markers demonstrates high sensitivity (84.9%) and specificity (83.3%) for colorectal cancer (CRC) detection, even at tumor DNA levels as low as 0.05%. In comparative studies, it outperformed the SEPT9 assay, particularly for early-stage CRC and precancerous lesions. In a longitudinal cohort, mqMSP identified ctDNA in 89% of preoperative cases and predicted recurrence with a median 8-month lead time over imaging. Its low cost and simplicity support its use for routine CRC monitoring post-surgery.
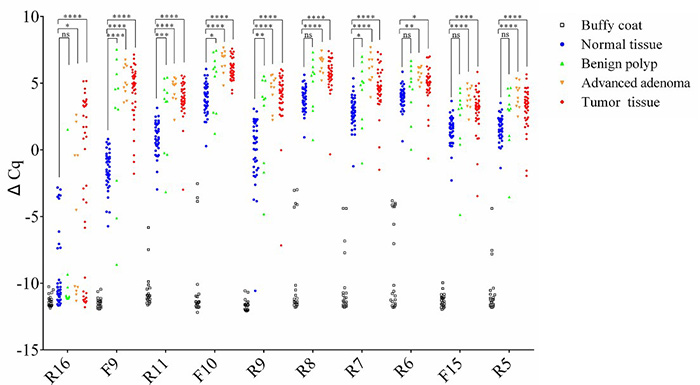
Figure 3. Selection and validation of multiple DNA methylation biomarkers within the SEPT9 gene. Samples from 40 pairs of CRCs (red) and surrounding normal tissues (blue), 10 advanced adenomas (orange), 10 benign polyps (green), and 20 buffy coats (black) were analyzed for the 10 selected markers. (Jin et al., 2020)
Customer Testimonials
-
“We needed a partner to develop a high-sensitivity assay for detecting histone methylation changes in response to novel epigenetic modulators. Creative BioMart delivered a robust, reproducible assay within weeks, helping us rapidly screen small-molecule libraries. Their technical team was responsive and highly knowledgeable throughout.”
— Head of Discovery Biology | Mid-Sized Biotech Firm
-
“Creative BioMart supported our development of a DNA methylation assay for early cancer biomarker discovery. They customized the workflow for bisulfite-treated DNA and incorporated NGS-based detection, significantly improving our throughput and data resolution.”
— Director of Translational Research | Oncology-Focused Pharma Company
-
“We required a multiplexed assay to simultaneously monitor histone acetylation and DNA methylation changes in stem cell models. Creative BioMart helped design a dual-readout platform tailored to our epigenetic screening program, saving us substantial time and internal resources.”
— Senior Scientist | Regenerative Medicine Startup
-
“We partnered with Creative BioMart to create a ChIP-based assay to track histone mark occupancy during drug treatment. The assay they delivered showed excellent signal-to-noise ratios and worked seamlessly with our automated liquid handlers. The data quality exceeded expectations.”
— Principal Investigator | Academic Medical Center
-
“Our team was exploring non-coding RNA regulators of chromatin state and needed a sensitive assay to track changes in repressive marks like H3K27me3. Creative BioMart proposed a tailored approach using antibody-based detection and quantitative PCR readouts—reliable, scalable, and publication-ready.”
— Epigenetics Group Lead | Global Pharmaceutical Company
FAQs
-
Q: What types of epigenetic modifications can your assays detect?
A: Our assays can detect a wide range of epigenetic modifications, including DNA methylation, histone modifications (such as acetylation, methylation, and phosphorylation), and non-coding RNA expression (such as miRNAs and lncRNAs). -
Q: Can you develop custom assays for specific research needs?
A: Yes, we specialize in developing custom assays tailored to meet the unique requirements of each client's research objectives. Our team works closely with clients to design and optimize assays that are optimized for sensitivity, specificity, and reproducibility. -
Q: What types of samples can your assays be applied to?
A: Our assays can be applied to a variety of biological samples, including tissues, cells, and biofluids (such as blood, plasma, and serum). We work with clients to validate and optimize our assays for use with their specific sample types. -
Q: How long does it take to develop and validate an assay?
A: The timeline for assay development and validation depends on the complexity of the project and the specific requirements of the client. Typically, the process can take anywhere from several weeks to a few months. We work closely with clients to provide regular updates and ensure that the project stays on schedule. -
Q: Do you provide data analysis and interpretation services?
A: Yes, we offer comprehensive data analysis and interpretation services as part of our assay development package. Our team of experts utilizes advanced bioinformatics tools and algorithms to process and analyze the complex datasets generated by our assays, providing clients with actionable insights for their research and development projects. -
Q: How can I get started with your services?
A: To get started, please contact us to schedule an initial consultation. Our team will discuss your research objectives and requirements in detail, and we will provide a customized proposal outlining the scope of the project, timeline, and costs.
Resources
Related Services
Related Products
References:
- Borges BDN. Epigenetic alterations in canine mammary cancer. Genet Mol Biol. 2022;45(3 suppl 1):e20220131. doi:10.1590/1678-4685-gmb-2022-0131
- Jin S, Zhu D, Shao F, et al. Efficient detection and post-surgical monitoring of colon cancer with a multi-marker DNA methylation liquid biopsy. Proc Natl Acad Sci USA. 2021;118(5):e2017421118. doi:10.1073/pnas.2017421118
- Porazzi P, Petruk S, Pagliaroli L, et al. Targeting chemotherapy to decondensed H3K27me3-marked chromatin of AML cells enhances leukemia suppression. Cancer Research. 2022;82(3):458-471. doi:10.1158/0008-5472.CAN-21-1297
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

