DNA methyltransferases (DNMTs)
Related Symbol Search List
Immunology Background
Background
DNA methyltransferases (DNMTs) are a group of enzymes that play a vital role in the epigenetic modification of DNA. Their primary function is to add a methyl group to the cytosine residue in DNA molecules, leading to the formation of 5-methylcytosine (5mC). DNA methylation is an essential epigenetic mechanism involved in gene regulation, genomic stability, and cellular differentiation.
Members of DNMTs
Here are some common DNMT members found in different species:
| Species | Members of DNMTs |
|---|---|
| Mammals |
DNMT1: Known as the maintenance methyltransferase, responsible for copying methylation patterns during DNA replication. Found in mammals including humans, mice, and other vertebrates. DNMT3A and DNMT3B: De novo methyltransferases that establish new methylation patterns. Present in mammals and play essential roles in development and cellular differentiation. DNMT3L: Functions as a regulatory factor that enhances the activity of DNMT3A and DNMT3B. Found in mammals, particularly important in germ cells. |
| Plants |
MET1 (Methyltransferase 1): The plant homolog of DNMT1 in mammals, responsible for maintaining DNA methylation patterns during DNA replication in plants. CMT3 (Chromomethylase 3): Involved in maintaining methylation in plant heterochromatin regions. DRM (Domains Rearranged Methyltransferase): Responsible for de novo methylation in plants and plays a role in regulating gene expression. |
| Bacteria | Bacterial DNA methyltransferases vary widely in structure and function. They are involved in various processes such as restriction-modification systems, bacterial defense mechanisms, and regulation of gene expression. |
| Other Species |
DNMTs in Invertebrates: Invertebrates like Drosophila melanogaster (fruit fly) and Caenorhabditis elegans (nematode worm) also possess DNMT homologs that participate in regulating gene expression and development. DNMTs in Fungi: Fungal species like Saccharomyces cerevisiae (baker's yeast) have DNMTs involved in epigenetic regulation and maintaining genome stability. |
These are just a few examples of DNMT members found in different species, each playing essential roles in establishing and maintaining DNA methylation patterns, regulating gene expression, and contributing to the epigenetic landscape of diverse organisms.
Structure Features and Functions of DNMT Members
| Members | Details |
|---|---|
| DNMT1 |
Maintenance Methyltransferase Structural Features: Contains regulatory domains and a catalytic domain. It includes an RFTS domain for replication foci targeting and a CXXC domain for DNA binding. Function: DNMT1 is primarily responsible for maintaining existing DNA methylation patterns during DNA replication. |
| DNMT3A and DNMT3B |
De Novo Methyltransferases Structural Features: Comprise regulatory and catalytic domains. Lack the RFTS domain found in DNMT1. Function: DNMT3A and DNMT3B are involved in establishing new DNA methylation patterns, particularly during early development and in response to environmental stimuli. |
| DNMT3L |
Regulatory Factor Structural Features: Lacks catalytic activity but interacts with DNMT3A and DNMT3B. Function: DNMT3L enhances the activity and specificity of DNMT3A and DNMT3B, playing a crucial role in modulating DNA methylation processes. |
Understanding the structure and function of DNA methyltransferases is vital for unraveling their roles in epigenetic regulation, disease processes, and potential therapeutic interventions targeting aberrant DNA methylation patterns.
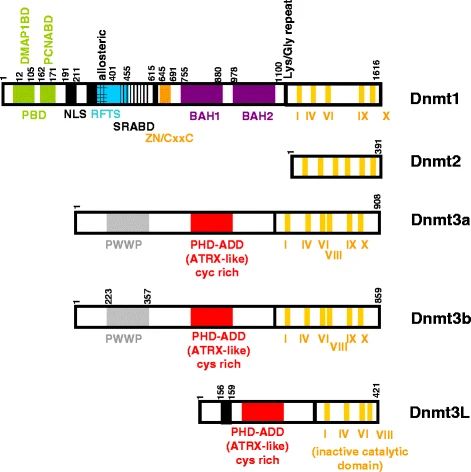 Fig.1 Representative structure of DNMTs. (Hervouet, E., et al., 2018)
Fig.1 Representative structure of DNMTs. (Hervouet, E., et al., 2018)Functions of DNMTs in Cells
DNMTs have crucial roles in various cellular processes and functions. Here are some key functions of DNMTs in cells:
| Functions | Details |
|---|---|
| DNA Methylation | The primary function of DNMTs is to add a methyl group to the cytosine residues in DNA, resulting in DNA methylation. This epigenetic modification plays a fundamental role in gene regulation by influencing gene expression patterns. DNMTs establish and maintain DNA methylation patterns, which can affect gene activity and cellular function. |
| Gene Silencing | DNMTs contribute to gene silencing by methylating specific regions of DNA, such as gene promoters or regulatory elements. DNA methylation in these regions can block the binding of transcription factors and other regulatory proteins, preventing gene transcription and effectively silencing gene expression. |
| Genomic Stability | DNMTs help maintain genomic stability by ensuring the faithful inheritance of DNA methylation patterns during DNA replication. DNMT1, in particular, is responsible for copying the DNA methylation patterns from the parent strand to the newly synthesized daughter strand, preserving epigenetic information. |
| Developmental Processes | DNMTs play critical roles in embryonic development and cellular differentiation. During early embryogenesis, DNMT3A and DNMT3B establish de novo DNA methylation patterns, which are necessary for proper development and cell lineage specification. DNA methylation changes, guided by DNMTs, contribute to the regulation of gene expression during tissue development and differentiation. |
| X-Chromosome Inactivation | DNMTs are involved in X-chromosome inactivation, a process that occurs in female mammals to balance gene expression between the sexes. DNMT3A and DNMT3B are responsible for initiating DNA methylation on one of the two X chromosomes in each cell, leading to its inactivation and gene silencing. |
| Imprinting | Imprinting refers to the differential DNA methylation of specific genes depending on their parental origin. DNMTs are involved in establishing and maintaining DNA methylation patterns at imprinted gene loci. Imprinting plays a significant role in controlling gene expression in a parent-of-origin-specific manner. |
| Transposon Silencing | Transposons are repetitive DNA elements with the ability to move within the genome. DNMTs help regulate transposons by adding methyl groups to their DNA sequences, which helps to silence and maintain their inactivity. This prevents the disruption of genomic integrity caused by transposon mobility. |
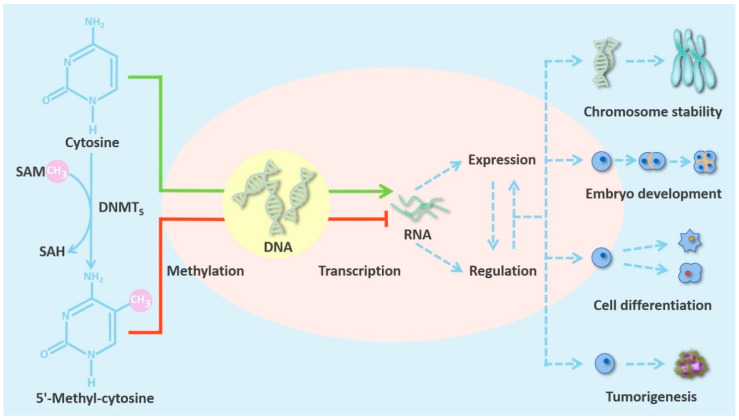 Fig.2 The characteristics of DNMTs in mammalian cells. DNMTs catalyze DNA methylation, which alters patterns of gene regulation and expression. This in turn affects chromosome stability, embryo development, and cell differentiation. Aberrant DNMTs can induce tumorigenesis. (Zhang J, et al., 2022)
Fig.2 The characteristics of DNMTs in mammalian cells. DNMTs catalyze DNA methylation, which alters patterns of gene regulation and expression. This in turn affects chromosome stability, embryo development, and cell differentiation. Aberrant DNMTs can induce tumorigenesis. (Zhang J, et al., 2022)The Impact of DNMTs on Disease
Cancer
One of the most well-studied examples of DNMTs in disease is in cancer. Aberrant DNA methylation patterns, often resulting from dysregulated DNMT activity, can lead to silencing of tumor suppressor genes or activation of oncogenes, contributing to the development and progression of various cancers. For instance, hypomethylation of oncogenes such as RAS and MYC has been associated with increased tumor growth and metastasis, while hypermethylation of tumor suppressor genes like p16 and MLH1 can lead to their inactivation and unchecked cell proliferation. Therapies targeting DNMTs, such as the DNMT inhibitors azacitidine and decitabine, have been developed to reverse these abnormal DNA methylation patterns and are currently used in the treatment of certain cancers.
Neurodevelopmental Disorders
In neurodevelopmental disorders, DNMT dysregulation has also been implicated in disease pathogenesis. For example, mutations in the DNMT3A gene have been found in individuals with autism spectrum disorder and intellectual disabilities, suggesting a role for DNMTs in regulating neuronal development and function. Additionally, altered DNA methylation patterns have been observed in patients with neurodegenerative diseases such as Alzheimer's and Parkinson's disease, indicating a potential link between DNMT activity and disease progression.
Autoimmune Diseases
In autoimmune diseases, aberrant DNA methylation patterns can lead to the misregulation of immune response genes, predisposing individuals to autoimmunity. For example, hypomethylation of genes involved in immune tolerance, such as FOXP3 in regulatory T cells, has been associated with the development of autoimmune diseases like systemic lupus erythematosus and rheumatoid arthritis. Targeting DNMTs with inhibitors has shown promise in modulating DNA methylation patterns and potentially reversing autoimmunity.
Cardiovascular Diseases
DNMTs have been implicated in various cardiovascular diseases through their role in modifying the expression of genes involved in cardiac function and vascular health. Abnormal DNA methylation patterns can influence the development of atherosclerosis, hypertension, and heart failure. For example, in atherosclerosis, increased levels of DNMT1 have been associated with enhanced methylation of genes that promote inflammation and vascular remodeling. The methylation of the IL-6 and TNF-α genes, which are involved in inflammatory responses, can lead to an increased risk of plaque formation in arteries.
Metabolic Disorders
DNMTs also play a significant role in metabolic diseases such as obesity, diabetes, and metabolic syndrome. They regulate the expression of genes involved in metabolic pathways, including insulin signaling, lipid metabolism, and inflammation. For instance, in type 2 diabetes, altered DNMT activity has been linked to abnormal methylation patterns of genes related to insulin resistance. For instance, hypermethylation of the PPARγ gene, which is crucial for adipocyte differentiation and lipid metabolism, can affect glucose homeostasis and contribute to the development of insulin resistance.
Infectious Diseases
Impact: DNMTs can be modulated during infectious diseases, influencing the host's immune response and pathogen interaction. Pathogens can exploit host DNMTs to evade the immune response or induce a state of immune tolerance. For instance, in viral infections, such as HIV, DNMTs can mediate the methylation of the host genome, silencing genes that are necessary for effective antiviral responses. In some cases, DNMT3B is upregulated in response to viral infection, leading to increased methylation of pro-inflammatory cytokine genes, thus impairing the immune response.
Aging-Related Diseases
Impact: DNMTs are involved in the aging process, influencing epigenetic changes that accumulate over time and contribute to age-related diseases, including neurodegeneration, cardiovascular diseases, and metabolic disorders. For example, in neurodegenerative diseases like Alzheimer's disease, altered DNMT expression and activity have been observed, leading to changes in the methylation patterns of genes associated with neuronal survival and synaptic function. For instance, hypermethylation of the gene encoding brain-derived neurotrophic factor (BDNF) has been implicated in cognitive decline and neurodegeneration.
Reproductive Disorders
DNMTs play a critical role in reproductive health, influencing genomic imprinting, fertility, and early embryonic development. Abnormal DNMT activity can lead to reproductive failures and developmental disorders. For example, in conditions like polycystic ovary syndrome (PCOS), altered DNMT expression has been linked to the improper regulation of genes involved in ovarian function and hormone signaling. Changes in DNA methylation can impact insulin sensitivity, which is often disrupted in PCOS patients.
Metastatic Diseases
Beyond cancer, DNMTs play a role in metastasis in various conditions, affecting cell migration and invasion through epigenetic regulation of epithelial-to-mesenchymal transition (EMT) processes. For example, in chronic lung diseases such as pulmonary fibrosis, dysregulated DNMT activity has been shown to promote EMT, enhancing the migratory capacity of fibroblasts and contributing to lung tissue remodeling and scarring.
The effects of DNMTs extend beyond cancer, neurodevelopmental disorders, and autoimmune diseases. Their regulatory roles in cardiovascular health, metabolic conditions, infectious diseases, aging, reproductive health, and other disorders underline the complexity of epigenetic influences in human diseases. Understanding these impacts can potentially lead to novel therapeutic interventions targeting DNMTs to mitigate disease progression and improve patient outcomes. Continued research in this area is critical for elucidating the full range of DNMT-related mechanisms in health and disease.
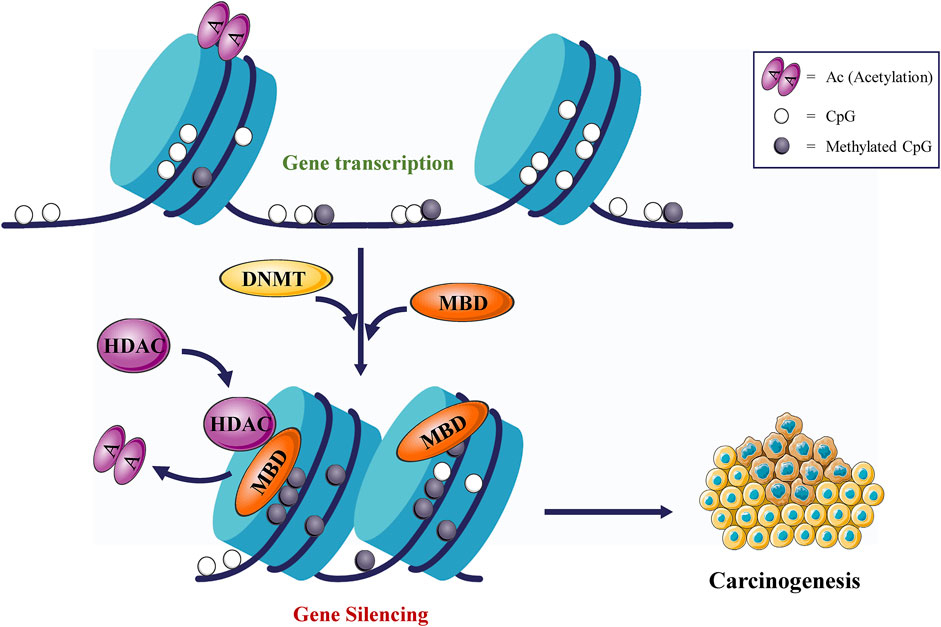 Fig.3 DNMTs cause gene silencing and promote tumorigenesis. (Liu P, et al., 2022)
Fig.3 DNMTs cause gene silencing and promote tumorigenesis. (Liu P, et al., 2022)Case Study
Case 1: Laranjeira ABA, Hollingshead MG, Nguyen D, Kinders RJ, Doroshow JH, Yang SX. DNA damage, demethylation, and anticancer activity of DNA methyltransferase (DNMT) inhibitors. Sci Rep. 2023;13(1):5964.
The impact of DNA damage and demethylation on the effectiveness of DNA methyltransferase inhibitors (DNMTi) in cancer treatment is a topic that requires further exploration. Recent research delved into the role of DNMT1 gene deletion or disruption (DNMT1-/-) in influencing the anticancer effects of a specific class of DNMT inhibitors in various settings, including laboratory studies, animal models, and human cancer cases.
The study revealed that the deletion of the DNMT1 gene significantly reduced the cytotoxic and growth-inhibitory effects of DNMT inhibitors such as decitabine, azacitidine, and 5-aza-4'-thio-2'-deoxycytidine (aza-T-dCyd) in colon and breast cancer cell lines. These drugs induced DNA damage alongside DNMT1 inhibition, leading to G2/M cell-cycle arrest, apoptosis, and increased expression of p21 in cells with intact DNMT1 compared to those with DNMT1 deletion, with aza-T-dCyd demonstrating the highest potency.
In animal models carrying HCT116 DNMT1+/+ xenograft and bladder patient-derived xenograft (PDX) tumors, tumor growth was notably inhibited, DNMT1 levels were reduced, and p21 expression was upregulated. Interestingly, DNMT1 gene deletions were observed in approximately 9% of human colon cancers and other cancer types to varying extents.
Decitabine and azacitidine were found to demethylate CDKN2A/CDKN2B genes regardless of the DNMT1 status, while increasing histone-H3 acetylation and re-expression of p16INK4A/p15INK4B specifically in DNMT1-/- conditions. This suggests that the absence of DNMT1 confers resistance to DNMT inhibitors, with the effectiveness of these drugs closely tied to their DNA damage effects. Patients with DNMT1 gene deletions may exhibit a reduced response to DNMT inhibitor treatments based on these findings.
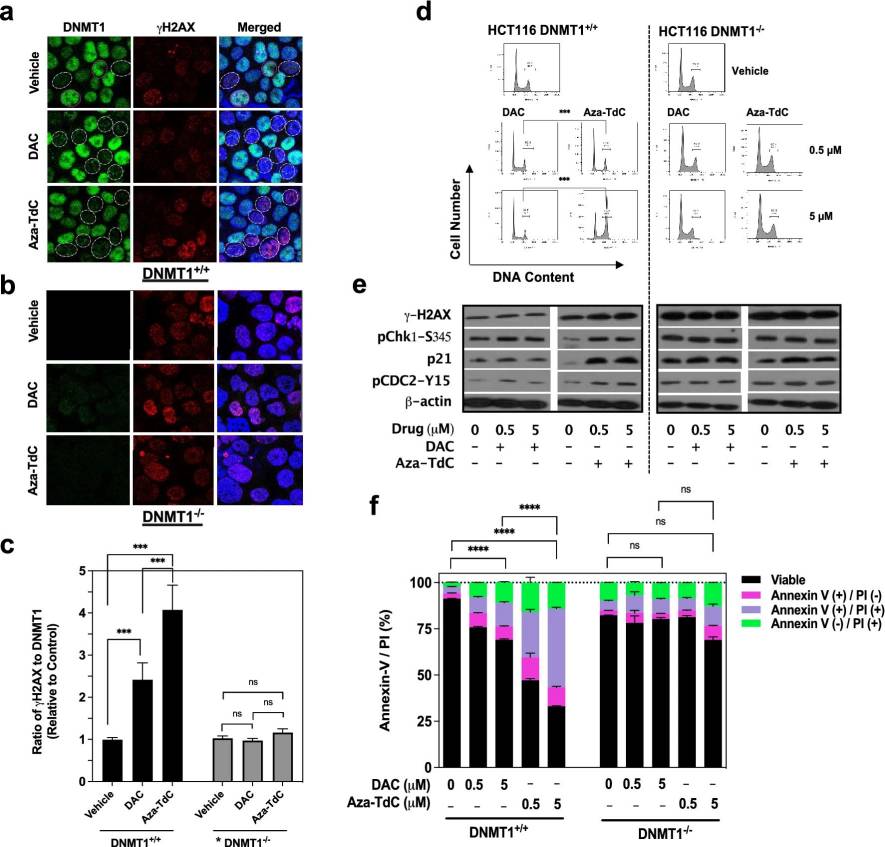 Fig.1 Effects of DNMTi on DNMT1 and DNA damage, cell-cycle, and apoptosis in HCT116 cells.
Fig.1 Effects of DNMTi on DNMT1 and DNA damage, cell-cycle, and apoptosis in HCT116 cells.Case 2: Yu J, Qin B, Moyer AM, et al. DNA methyltransferase expression in triple-negative breast cancer predicts sensitivity to decitabine. J Clin Invest. 2018;128(6):2376-2388.
In the context of Triple-negative breast cancer (TNBC), a challenging disease with limited targeted treatments, researchers are exploring the potential of DNA methyltransferase (DNMT) inhibitors like 5-aza-2'-deoxycytidine (decitabine) as a therapeutic strategy. While decitabine has shown effectiveness in hematological neoplasms by reversing DNA hypermethylation-induced silencing of tumor suppressors, its efficacy in solid tumors, including TNBC, varies significantly. To address this, efforts have been focused on identifying predictive biomarkers for decitabine sensitivity in TNBC.
A recent study investigated the correlation between DNMT protein levels and response to decitabine in patient-derived xenograft (PDX) organoids derived from both chemotherapy-sensitive and -resistant TNBCs. The findings suggested that DNMT levels could potentially serve as biomarkers for predicting response to decitabine treatment. Additionally, the study revealed that long-term, low-concentration decitabine treatment led to the degradation of all three DNMTs (DNMT1, DNMT3A, and DNMT3B) both in laboratory settings and in animal models.
Further investigation uncovered a mechanism wherein the E3 ligase TNF receptor–associated factor 6 (TRAF6) facilitated the ubiquitination of DNMT proteins, subsequently targeting them for lysosome-dependent degradation. Inhibition of TRAF6 resulted in resistance to decitabine-induced DNMT degradation, highlighting the role of TRAF6 in regulating DNMT protein levels and potentially influencing responses to DNMT inhibitors in TNBC. This study provides insights into a potential mechanism for modulating DNMT protein degradation and underscores the importance of DNMT levels as potential biomarkers for predicting responses to DNMT inhibitors in TNBC therapy.
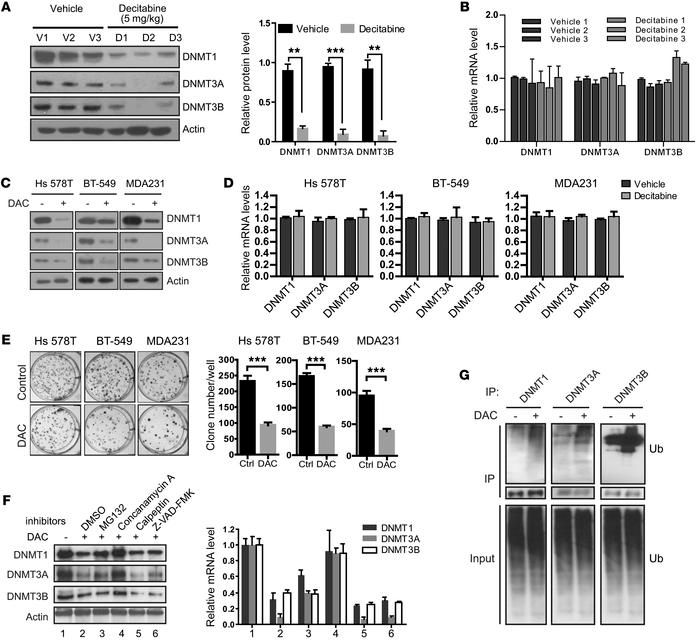 Fig.2 Decitabine treatment induced lysosome-dependent degradation of DNMTs.
Fig.2 Decitabine treatment induced lysosome-dependent degradation of DNMTs.Related References
- Hervouet, E., Peixoto, P., Delage-Mourroux, R. et al. Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma. Clin Epigenet 10, 17 (2018).
- Liu P, Yang F, Zhang L, et al. Emerging role of different DNA methyltransferases in the pathogenesis of cancer. Front Pharmacol. 2022;13:958146.
- Zhang J, Yang C, Wu C, Cui W, Wang L. DNA methyltransferases in cancer: biology, paradox, aberrations, and targeted therapy. Cancers (Basel). 2020;12(8):2123.

