Chromatin Remodelers
Creative BioMart Chromatin Remodelers Product List
Immunology Background
Background
Chromatin remodelers are essential and dynamic molecular machines that regulate the structure and accessibility of chromatin, the complex of DNA and proteins that make up chromosomes within a cell's nucleus. These remodelers play a crucial role in controlling gene expression, DNA replication, repair, and recombination by altering the interactions between DNA and histones, the proteins around which DNA is wrapped.
Unlike histone-modifying enzymes that add or remove chemical groups to histone proteins, chromatin remodelers use the energy derived from ATP hydrolysis to reposition, evict, or alter the composition of nucleosomes—the fundamental units of chromatin—along the DNA strand. By doing so, chromatin remodelers can regulate the accessibility of DNA to transcription factors and other regulatory proteins, thereby influencing gene expression patterns.
There are several families of chromatin remodelers, each with distinct structural features and functional roles. These remodelers can be broadly categorized into four main classes based on their structural domains and mechanisms of action: SWI/SNF (switch/sucrose non-fermentable), ISWI (imitation switch), CHD (chromodomain helicase DNA-binding), and INO80 (inositol requiring 80). SNF2-family ATPase domain present in all remodeler families is split into 2 parts; DExx and HELIC. The ATPase domain of the SWI/SNF, ISWI, and CHD remodeler families possesses a short insertion, whereas remodelers of the INO80 family consist of a long insertion. The exclusive domains reside adjacent to the ATPase domain. SWI/SNF remodelers contain bromodomains; ISWI remodelers - SANT-SLIDE modules; CHD remodelers - tandem chromodomains and members of the INO80 family possess HAS (helicase SANT) domains. Each of these domains plays a role in remodeler recruitment to chromatin or interacting with specific histone modifications and/or they are involved in the regulation of the ATPase activity of the remodeler.
Chromatin remodelers are involved in a wide range of cellular processes, including development, differentiation, and responses to environmental cues. Dysregulation of chromatin remodeling processes has been implicated in various diseases, including cancer, neurological disorders, and developmental abnormalities.
In summary, chromatin remodelers are integral components of the epigenetic machinery that governs gene expression by modulating the structure and accessibility of chromatin. Their intricate functions highlight their significance in cellular processes and underscore their potential as therapeutic targets for various diseases.
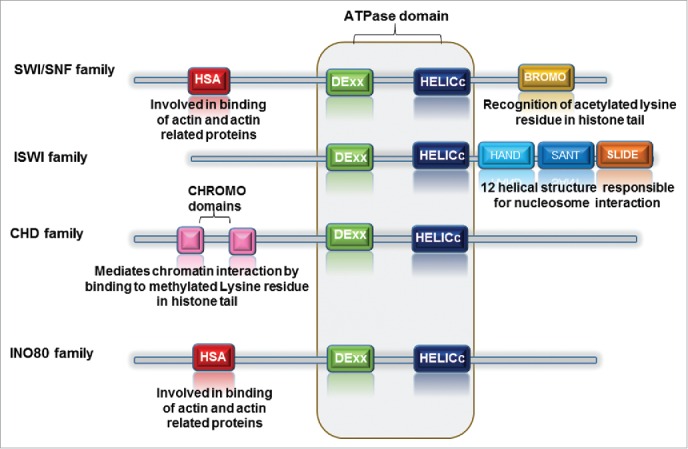 Fig.1 Diagrammatic representation of chromatin remodeler family highlighting the conserved domain with each family member. (Tyagi M, et al., 2016)
Fig.1 Diagrammatic representation of chromatin remodeler family highlighting the conserved domain with each family member. (Tyagi M, et al., 2016)
The DEX and HELICc domains are conserved throughout the family. However, HAND, SANT, and SLIDE domains are specific to ISWI family, whereas BROMO domain distinguishes the SWI/SNF family. The presence of CHROMO domains is characteristic of CHD family.
Table 1. Four chromatin-remodeler families, subunits, and their respective functional domains. (Cui G, et al., 2022)
| Families | Subunits | Domains | Functions |
|---|---|---|---|
| SWI/SNF | SWI1, SNF11, Swp82, SWI2/SNF2, Swp73, SWI3, Arp9, SNF5, Arp7, SNF6, Swp29 | HSA | Actin binding |
| DExx | ATPase | ||
| HELICc | ATPase | ||
| BROMO | Acetylated lysine binding | ||
| ISWI | ISW1, P74, p110, p105, ACF, RSF, CERF, CHRAC, NURF, NoRC, WICH, b-WICH | DExx | ATPase |
| HELICc | ATPase | ||
| SANT | Histone binding | ||
| SLIDE | Histone binding | ||
| CHD | CHD1, CHD2, CHD3, CHD4, CHD9, NuRD | CHROMO | Histone binding |
| DExx | ATPase | ||
| HELICc | ATPase | ||
| INO80 | Arp8, Arp4, Taf14, Ies4, Actin, Ino80, RvB1/2, Ies3, Ies1, Ies2, Arp5, Nhp10, Ies5, Ies6, SRCAP | HSA | Actin binding |
| DExx | ATPase | ||
| HELICc | ATPase |
Table 2. The major chromatin remodeler readers/domains and their binding histone targets. (Cui G, et al., 2022)
| Chromatin remodeler readers/domains | Histone target(s) | Example |
|---|---|---|
| Bromodomain | Acetylation Lysine | H3K14ac [50] |
| Chromo domain | Methylation Lysine | H3K4me, H3K9me, H3K27me, H3K36me |
| PWWP (Pro-Trp-Trp-Pro motif) | Methylation Lysine | H3K36me |
| PHD finger | Methylation Lysine | H3K4me |
| MBT (malignant brain tumor) repeat | Methylation Lysine | H4K20m |
| SANT domain | non-modified histone tails |
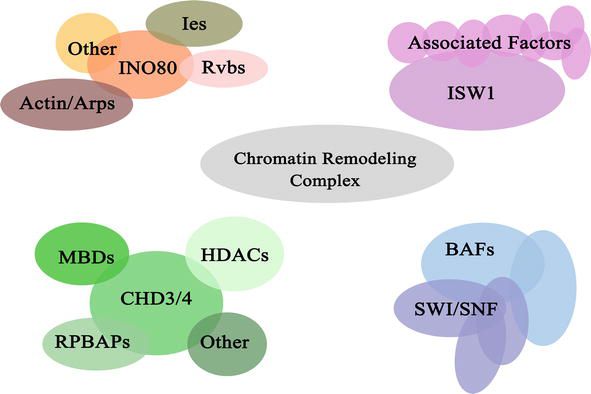 Fig.2 Composition of the chromatin remodeler complexes. (Cui G, et al., 2022)
Fig.2 Composition of the chromatin remodeler complexes. (Cui G, et al., 2022)Mechanisms of Chromatin Remodeling
Chromatin remodeling orchestrates a dynamic shift in chromatin structure, toggling between a condensed form (heterochromatin) and a relaxed state (euchromatin). This process involves two main mechanisms:
1. Covalent Modification of Chromatin Components
This includes epigenetic alterations to histone proteins such as acetylation, methylation, phosphorylation, and ubiquitination. Enzymes like histone acetyltransferase (HAT), histone deacetylase (HDAC), histone methyltransferase (HMT), histone demethylase (HDM), phosphokinase (PK), and phosphatase (PP) govern these modifications. DNA methylation is regulated by DNA methyltransferase (DNMT) and DNA demethylase.
2. Noncovalent Modifications by ATP-Dependent Chromatin Remodeling Complexes (CRCs)
This category includes families like SWI/SNF, ISWI, CHD, and INO80. Each family exhibits distinct domain structures. They share a common ATPase domain comprising DEXDc and HELICc domains, enabling them to utilize ATP hydrolysis energy to shift nucleosomes. Different members of these families possess unique auxiliary domains and employ diverse strategies for nucleosome repositioning.
In essence, chromatin remodeling is a sophisticated process governed by a balance of covalent modifications to histones and DNA, as well as noncovalent alterations facilitated by ATP-dependent chromatin remodelers. These mechanisms intricately regulate chromatin structure, impacting gene expression, cellular functions, and ultimately, cellular identity.
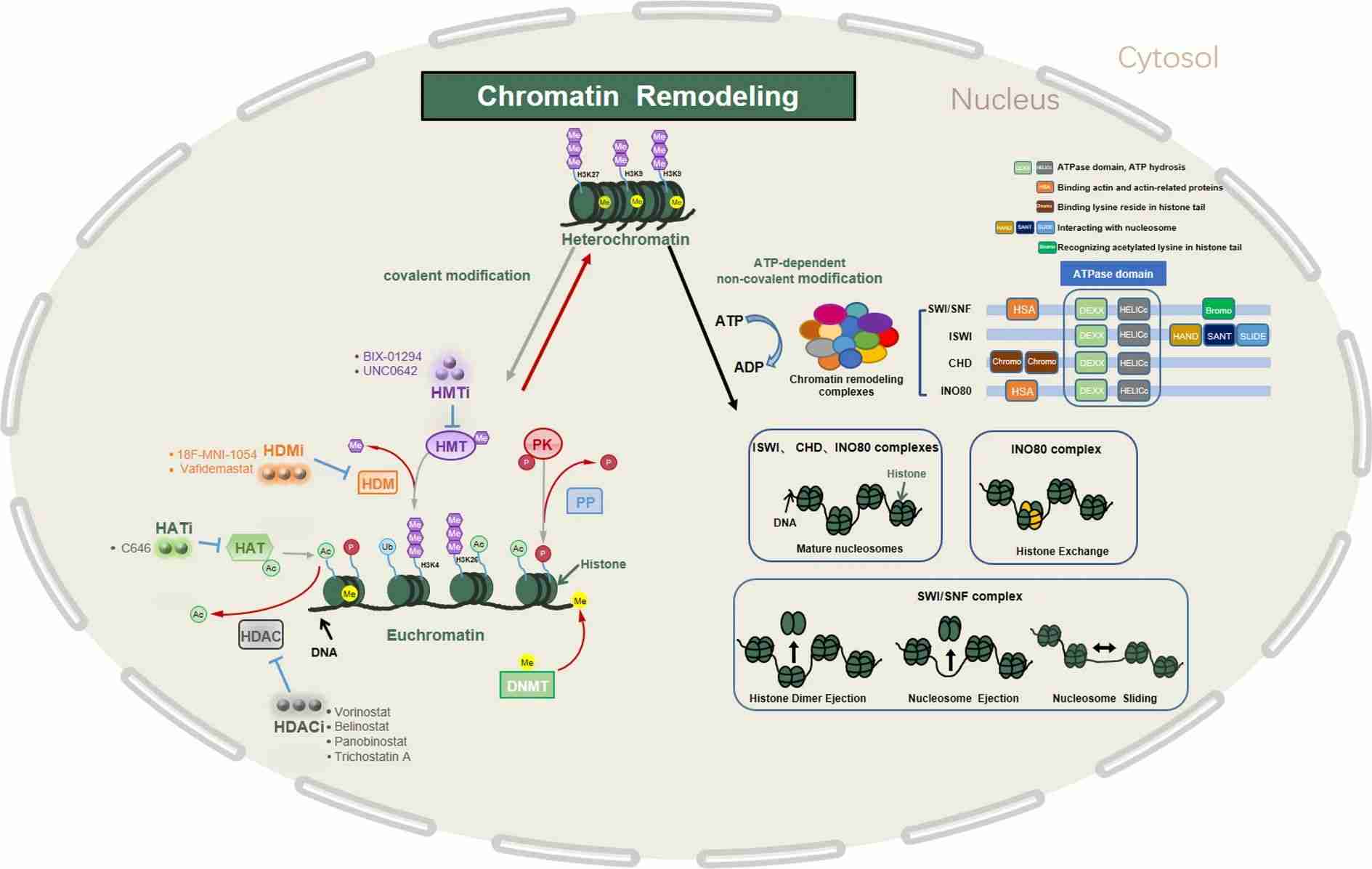 Fig.3 Summary of regulatory mechanisms of chromatin remodeling. (Jiang D, et al., 2023)
Fig.3 Summary of regulatory mechanisms of chromatin remodeling. (Jiang D, et al., 2023)Physiologic Functions of Chromatin Remodelers
| Functions | Details |
|---|---|
| Cell Differentiation |
Embryonic Development: During embryogenesis, chromatin remodelers participate in shaping the chromatin landscape to regulate the expression of genes crucial for cell fate determination and tissue differentiation. Cell Fate Specification: Chromatin remodeling is instrumental in establishing and maintaining cell identity by controlling the accessibility of specific genes that drive differentiation into specialized cell types. Stem Cell Maintenance: Chromatin remodelers help maintain the balance between self-renewal and differentiation in stem cells by modulating the chromatin structure at key regulatory regions. |
| Transcriptional Regulation |
Gene Expression: Chromatin remodelers control the accessibility of DNA to transcription factors and the transcriptional machinery, thus influencing the activation or repression of gene expression. Enhancer-Promoter Interactions: They facilitate the dynamic interactions between enhancers and promoters by modulating chromatin structure, allowing for precise control of gene expression patterns. Response to External Stimuli: Chromatin remodelers respond to external signals and environmental cues to regulate gene expression changes required for cellular adaptation and homeostasis. |
| Epigenetic Inheritance |
Memory of Gene Expression Patterns: Chromatin remodelers help establish and maintain epigenetic marks that can be inherited through cell divisions, ensuring the faithful transmission of gene expression patterns to daughter cells. Transgenerational Inheritance: They can also contribute to transgenerational epigenetic inheritance by influencing chromatin states that persist across multiple generations. |
Chromatin remodelers play pivotal roles in various physiological functions within cells, exerting their influence on critical processes like cell differentiation and transcriptional regulation. Here's a breakdown of how chromatin remodelers contribute to these essential functions:
The Role of Chromatin Remodelers in Disease Development and Treatment
Chromatin remodelers play a crucial role in the development and progression of various diseases by influencing gene expression patterns and chromatin structure. Here's an overview of how chromatin remodelers contribute to disease pathogenesis:
Cancer
- Tumorigenesis: Dysregulation of chromatin remodelers can lead to alterations in gene expression that promote uncontrolled cell growth and division, a hallmark of cancer development.
- Metastasis: Aberrant chromatin remodeling can drive the metastatic spread of cancer cells by regulating the expression of genes involved in invasion and migration.
- Therapeutic Resistance: Changes in chromatin structure mediated by remodelers can confer resistance to cancer therapies, making treatment less effective.
- Targeted Therapy: Utilizing chromatin remodelers as targets for cancer therapy can help modulate gene expression patterns in cancer cells. For example, inhibitors of specific chromatin remodelers like SWI/SNF complexes have shown promise in treating certain types of cancers, such as rhabdoid tumors.
- Reversal of Epigenetic Silencing: Chromatin remodelers can be used to reverse epigenetic silencing of tumor suppressor genes in cancer cells. Drugs targeting histone deacetylases (HDACs) and DNA methyltransferases (DNMTs) have been explored in various cancers, including hematological malignancies.
Neurological Disorders
- Neurodevelopmental Disorders: Altered chromatin remodeling in the brain can disrupt the expression of genes critical for brain development, contributing to neurodevelopmental disorders such as autism spectrum disorders and intellectual disabilities.
- Neurodegenerative Diseases: Dysregulation of chromatin remodelers can impact the expression of genes involved in neuronal function and survival, contributing to the pathogenesis of neurodegenerative conditions like Alzheimer's disease and Parkinson's disease.
- Epigenetic Modulation: Epigenetic modulation through targeting chromatin remodelers shows promise in treating neurodevelopmental disorders like Rett syndrome and neurodegenerative diseases such as Alzheimer's Disease. By modulating gene expression patterns and correcting aberrant gene expression, these approaches may offer novel therapeutic strategies for these conditions.
Immunological Disorders
- Autoimmune Diseases: Dysregulated chromatin remodeling can lead to abnormal gene expression in immune cells, triggering autoimmune responses against healthy tissues.
- Inflammatory Conditions: Chromatin remodelers can modulate the expression of pro-inflammatory genes, contributing to chronic inflammatory diseases like rheumatoid arthritis and inflammatory bowel disease.
- Modulating the Chromatin Landscape: Chromatin remodelers can be targeted to modulate the expression of immune-related genes in autoimmune diseases like lupus or rheumatoid arthritis, as well as in inflammatory conditions such as psoriasis or inflammatory bowel disease. Modulating the chromatin landscape may help attenuate aberrant immune responses and provide new treatment options for these conditions.
Metabolic Disorders
- Obesity and Diabetes: Altered chromatin remodeling can affect the expression of genes involved in metabolism and energy balance, potentially contributing to obesity and type 2 diabetes.
- Cardiovascular Diseases: Dysregulation of chromatin remodelers in cardiovascular cells can impact gene expression patterns related to heart function and vascular health, contributing to cardiovascular diseases like atherosclerosis and heart failure.
- Modulating Chromatin Structure: Targeting chromatin remodelers involved in metabolic gene regulation may offer potential treatments for metabolic disorders like type 2 diabetes and obesity. Modulating chromatin structure in metabolic tissues could impact gene expression related to glucose metabolism and adipogenesis.
Epigenetic Therapy
- Epigenetic Drugs: Targeting chromatin remodelers with epigenetic drugs can be a promising therapeutic strategy for certain diseases, including cancers where specific remodelers are dysregulated.
- Precision Medicine: Understanding the role of chromatin remodelers in disease pathogenesis can aid in the development of targeted therapies that modulate chromatin structure to reverse disease-associated gene expression changes.
- Combination Therapies: Combinations of chromatin remodeler inhibitors with other targeted therapies or immunotherapies can enhance treatment efficacy in various cancers. For example, combining HDAC inhibitors with immune checkpoint inhibitors in certain cancers has shown synergistic effects.
- Precision Medicine: Tailoring treatments based on the epigenetic profile of tumors, including chromatin remodeling patterns, can lead to more personalized and effective therapies. Precision medicine approaches using epigenetic drugs are being explored in oncology for improved patient outcomes.
By influencing gene expression and chromatin structure, chromatin remodelers emerge as key players in the development and progression of a wide range of diseases. Targeting these remodelers presents a promising avenue for therapeutic interventions and precision medicine approaches in various pathological conditions.
Case Study
Case 1: Prozzillo Y, Santopietro MV, Messina G, Dimitri P. Unconventional roles of chromatin remodelers and long non-coding RNAs in cell division. Cell Mol Life Sci. 2023;80(12):365. Published 2023 Nov 20. doi:10.1007/s00018-023-04949-8
Recent studies have investigated the localization and roles of various subunits of human chromatin remodeling complexes SRCAP and p400/TIP60 during cell division, particularly at the mitotic apparatus (centrosomes, spindle, and midbody) in HeLa and U2OS cell lines. Depletion of these subunits through RNA interference led to cell division defects in both mitosis and cytokinesis, indicating a potential direct involvement of chromatin remodeling subunits in cytokinesis.
Co-immunoprecipitation experiments conducted on chromatin-free protein extracts from telophase-synchronized HeLa cells supported the idea of a direct role of chromatin remodeling subunits in cytokinesis. During telophase, subunits such as SRCAP, BAF53a, and TIP60 interacted with proteins like α-tubulin, Aurora B, CIT-K, and other cytokinesis regulators associated with the midbody, suggesting their active participation in this process.
Interestingly, similar relocation behaviors during mitosis and meiosis were observed in the subunits of the DOM/TIP60 complex in Drosophila melanogaster, which are orthologous to those found in the human SRCAP and p400/TIP60 complexes. Given that the divergence between Drosophila and humans occurred around 780 million years ago, these findings strongly indicate that the recruitment of chromatin remodelers to the mitotic apparatus is an evolutionarily conserved and biologically functional process.
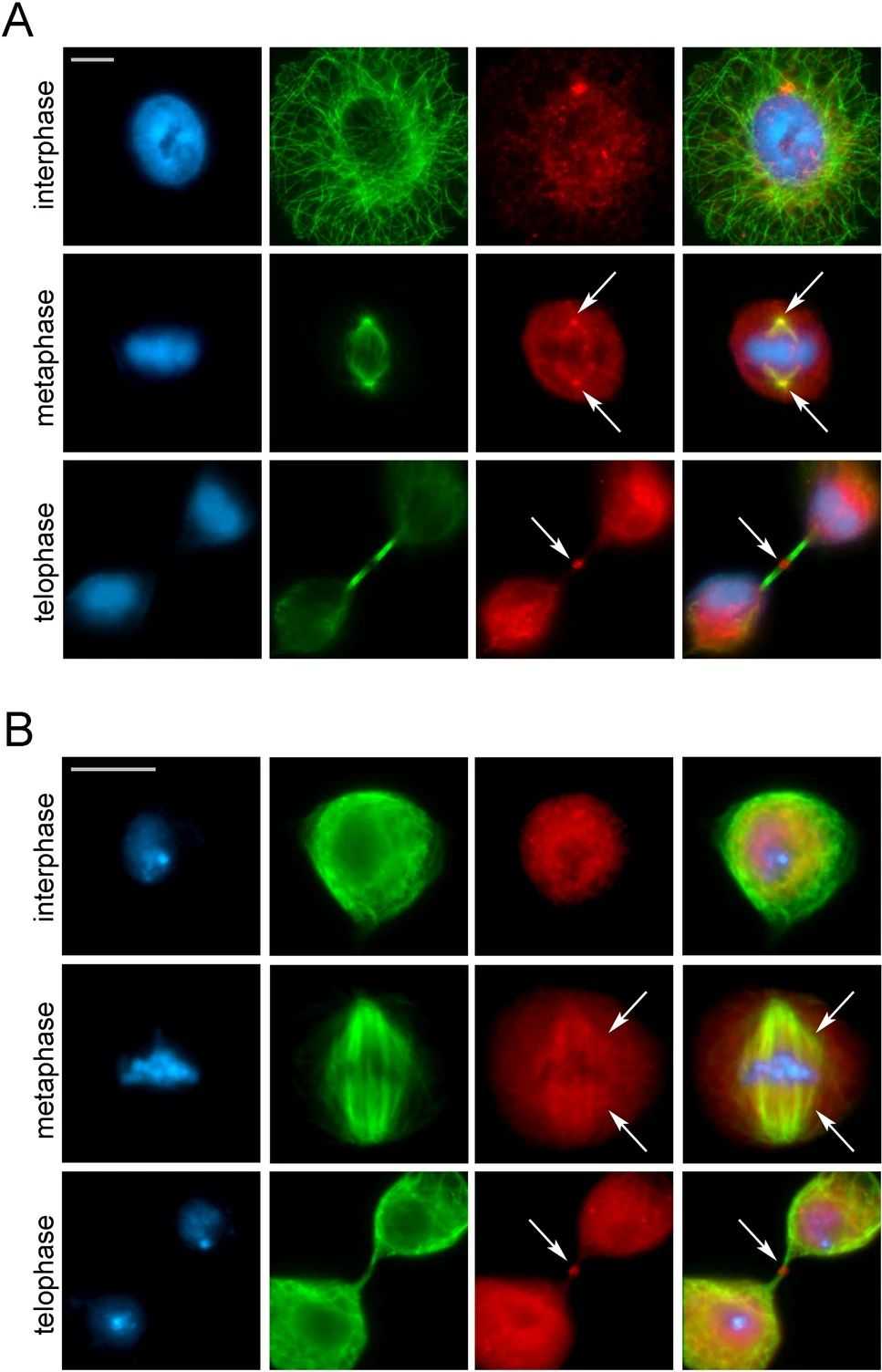 Fig.1 Evolutionarily conserved relocation of chromatin remodelers to the cell division apparatus.
Fig.1 Evolutionarily conserved relocation of chromatin remodelers to the cell division apparatus.Case 2: Pagliaroli L, Porazzi P, Curtis AT, et al. Inability to switch from ARID1A-BAF to ARID1B-BAF impairs exit from pluripotency and commitment towards neural crest formation in ARID1B-related neurodevelopmental disorders. Nat Commun. 2021;12(1):6469.
During development, subunit switches within the BAF chromatin remodeler play a critical role. The genes ARID1B and its paralog ARID1A encode mutually exclusive BAF subunits. De novo mutations causing haploinsufficiency in ARID1B are linked to neurodevelopmental disorders like Coffin-Siris syndrome, characterized by neurological and craniofacial abnormalities.
Using patient-derived induced pluripotent stem cells (iPSCs) from individuals with ARID1B haploinsufficiency and Coffin-Siris syndrome, researchers studied cranial neural crest cell (CNCC) formation. They found that ARID1B is active specifically during the initial stage of CNCC formation, coinciding with neuroectoderm specification, where it is part of a lineage-specific BAF configuration (ARID1B-BAF). ARID1B-BAF plays a crucial role in regulating pluripotency exit and lineage commitment by dampening the activity of numerous enhancers and genes within the NANOG and SOX2 networks. In iPSCs, these enhancers remain active due to ARID1A-containing BAF.
As differentiation begins, cells shift from ARID1A-containing BAF to ARID1B-BAF, leading to the suppression of the NANOG/SOX2 networks and the initiation of pluripotency exit. However, in Coffin-Siris patient cells, the switch from ARID1A to ARID1B-BAF does not occur, and ARID1A-BAF remains active at pluripotency enhancers throughout CNCC formation stages. This sustained NANOG/SOX2 activity hinders proper CNCC formation. Despite displaying typical neural crest cell markers (TFAP2A/SOX9-positive), ARID1B-haploinsufficient CNCCs also exhibit abnormal NANOG expression.
These findings suggest a link between ARID1B mutations, neuroectoderm specification, and a potential pathological mechanism underlying Coffin-Siris syndrome. The failure to undergo the ARID1A/ARID1B subunit switch disrupts normal developmental processes, impacting pluripotency regulation and lineage commitment, ultimately contributing to the manifestations of the syndrome.
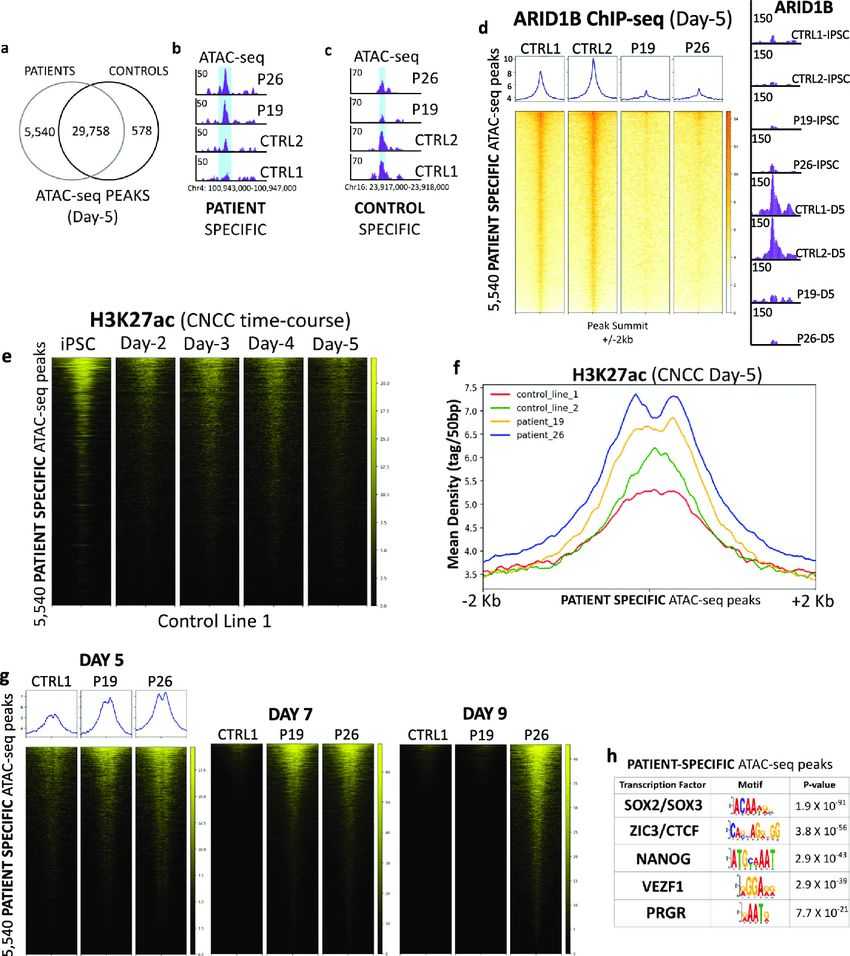 Fig.2 Chromatin remodeling at pluripotency enhancers is dysregulated in the patient cells.
Fig.2 Chromatin remodeling at pluripotency enhancers is dysregulated in the patient cells.Related References
- Tyagi M, Imam N, Verma K, Patel AK. Chromatin remodelers: We are the drivers!. Nucleus. 2016;7(4):388-404.
- Jiang D, Li T, Guo C, Tang TS, Liu H. Small molecule modulators of chromatin remodeling: from neurodevelopment to neurodegeneration. Cell Biosci. 2023;13(1):10.
- Cui G, Dong Q, Gai K, et al. Chromatin dynamics: Chromatin remodeler, epigenetic modification and diseases. Epigenetics-Regulation and New Perspectives, 2022.

