Histones
Creative BioMart Histones Product List
Immunology Background
Background
Histones are a group of proteins that play a fundamental role in organizing and packaging DNA in the cell nucleus. They are highly conserved and are found in eukaryotic organisms, including humans. The primary function of histones is to facilitate the compaction of DNA into a highly condensed and organized structure called chromatin, which is necessary for the efficient storage, replication, and expression of genetic information.
A nucleosome is a fundamental unit of chromatin composed of 147 base pairs of DNA and an octamer of core histone proteins, two copies each of histones H2A, H2B, H3, and H4. Histone proteins are basic and conserved from archaea to humans based on the amino acid sequence. Histones are typically small, positively charged proteins that interact with the negatively charged DNA molecule through electrostatic interactions. There are five main types of histones: H1, H2A, H2B, H3, and H4. These histones come together to form an octamer known as the nucleosome, which is the basic unit of chromatin.
The nucleosome consists of two copies each of histones H2A, H2B, H3, and H4, forming a protein core around which DNA is wrapped. The DNA helix makes approximately 1.7 turns around the histone octamer, resulting in about 147 base pairs of DNA being associated with each nucleosome. The linker histone H1 binds to the DNA between nucleosomes, helping to further compact and stabilize the chromatin structure.
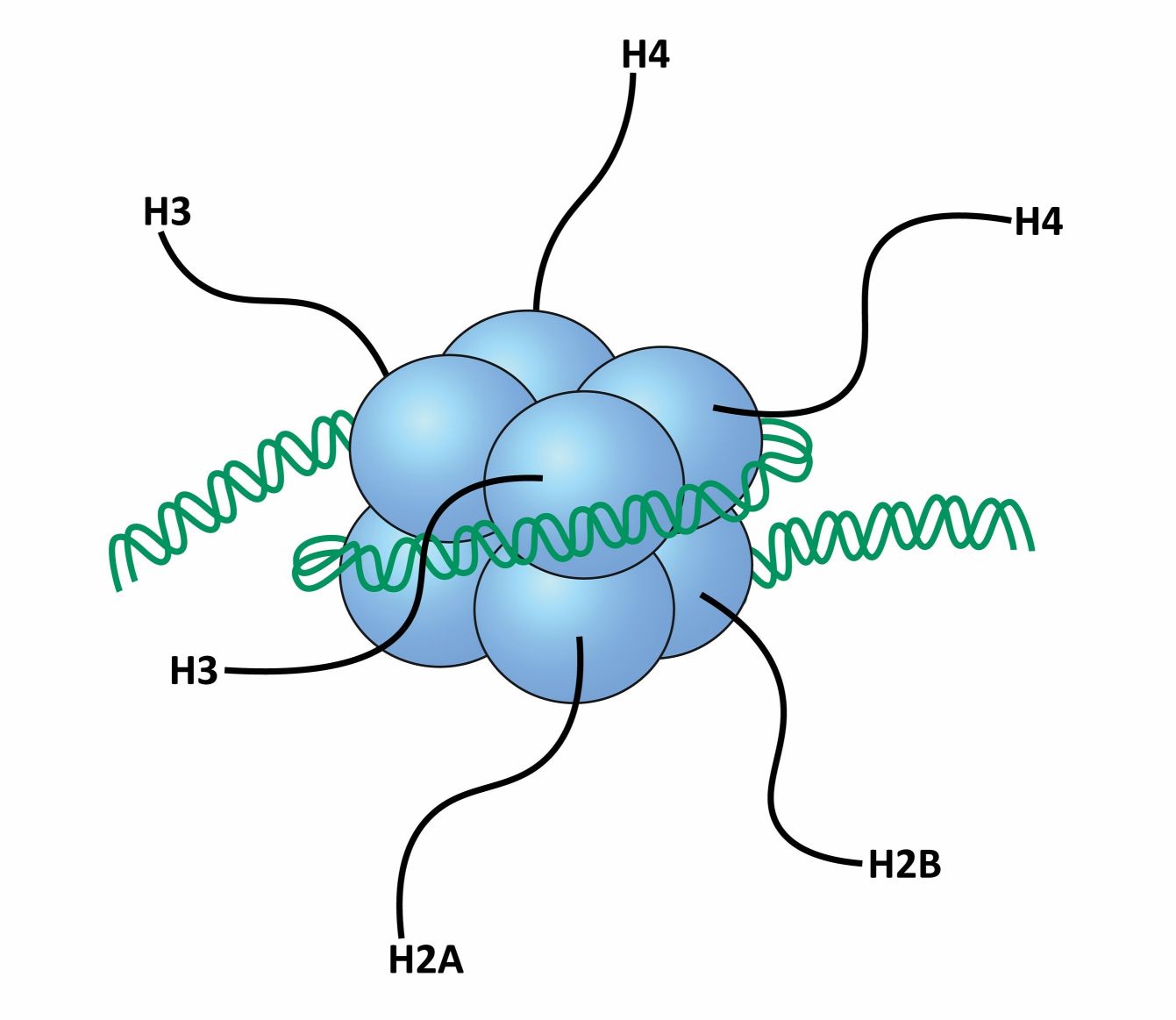
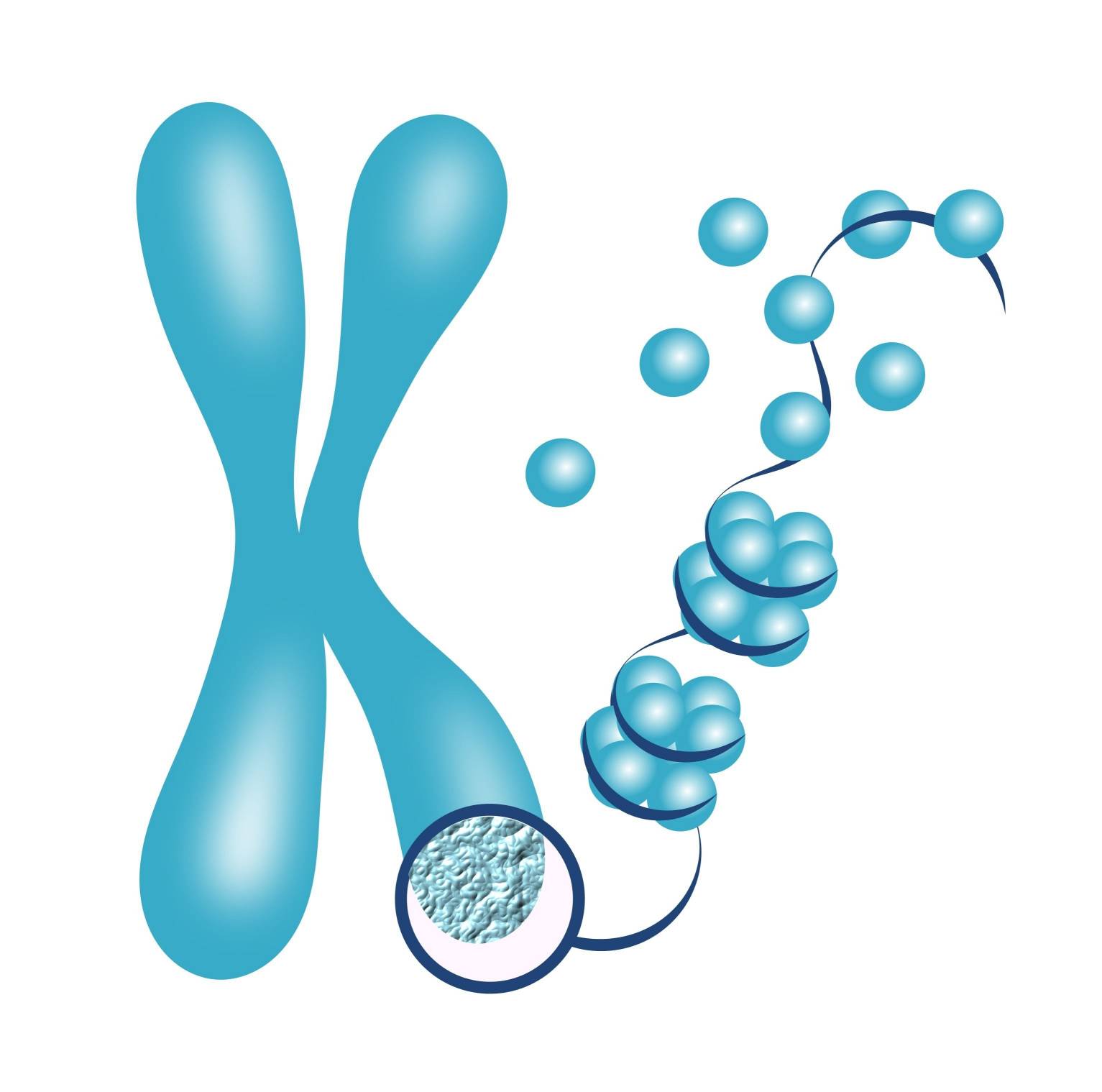
The assembly of nucleosomes along the DNA forms a "beads-on-a-string" structure, with the nucleosomes acting as the beads and the DNA linker regions connecting them. This structure can be further folded and condensed into higher-order structures, ultimately leading to the formation of chromosomes during cell division.
The interaction between histones and DNA is crucial for the compaction and organization of DNA within the nucleus. The positively charged amino acids in histones interact with the negatively charged phosphate groups of DNA, allowing histones to wrap and package the DNA molecule. This wrapping of DNA around the histone octamer not only compacts the genetic material but also protects it from damage and provides a mechanism for regulating gene expression.
The organization of DNA into chromatin and chromosomes is dynamic and can be modulated through various mechanisms, including histone modifications, nucleosome remodeling, and higher-order chromatin folding. These processes play important roles in regulating gene expression, DNA replication, DNA repair, and chromosome segregation during cell division.
In summary, histones are proteins that play a critical role in packaging DNA into a compact and organized structure within the cell nucleus. They form nucleosomes, the basic building blocks of chromatin, which help in DNA compaction and protect the genetic material. The interaction between histones and DNA is essential for the assembly of chromosomes and the regulation of gene expression.
Histone Variants
Histone variants are protein isoforms of canonical histones that differ in their amino acid sequence and structural properties. These variants are incorporated into chromatin in a replication-independent manner and can contribute to diverse chromatin functions. Here are some examples of histone variants and their roles:
| Variants | Functions |
|---|---|
| H2A Variants |
H2A.Z: H2A.Z is involved in transcriptional regulation and chromatin remodeling. It is often associated with gene promoters and enhancers, playing a role in regulating gene expression and facilitating nucleosome turnover. MacroH2A: MacroH2A is characterized by an extended C-terminal macrodomain. It is involved in transcriptional repression, X chromosome inactivation, and maintenance of genomic stability. |
| H2B Variants |
H2B.Z: Similar to H2A.Z, H2B.Z is associated with transcriptional regulation and chromatin remodeling. It affects nucleosome stability, gene expression, and DNA repair processes. H2B.W: H2B.W is implicated in germ cell-specific chromatin remodeling and spermatogenesis. |
| H3 Variants |
H3.3: H3.3 is involved in transcriptional regulation, DNA repair, and maintenance of genome integrity. It is often found in actively transcribed genes, enhancers, and regulatory regions. CENP-A: CENP-A replaces canonical H3 in centromeric chromatin, forming the specialized nucleosomes that play a crucial role in kinetochore assembly and proper chromosome segregation during cell division. |
| H4 Variants |
H4.2: H4.2 is a replication-dependent variant and is enriched in actively transcribed genes. It is involved in nucleosome stability and gene expression regulation. H4K20me1-enriched variants: These variants, such as H4K20me1-enriched H4 variants (H4K20me1-H4), are associated with heterochromatin formation and maintenance of genome stability. |
Histone variants contribute to the functional diversity of chromatin by influencing nucleosome structure, stability, and interactions with other proteins. They can regulate gene expression, chromatin remodeling, DNA repair, and various cellular processes. The presence of specific histone variants at specific genomic regions contributes to the establishment and maintenance of distinct chromatin states, ultimately influencing cellular functions and developmental processes.
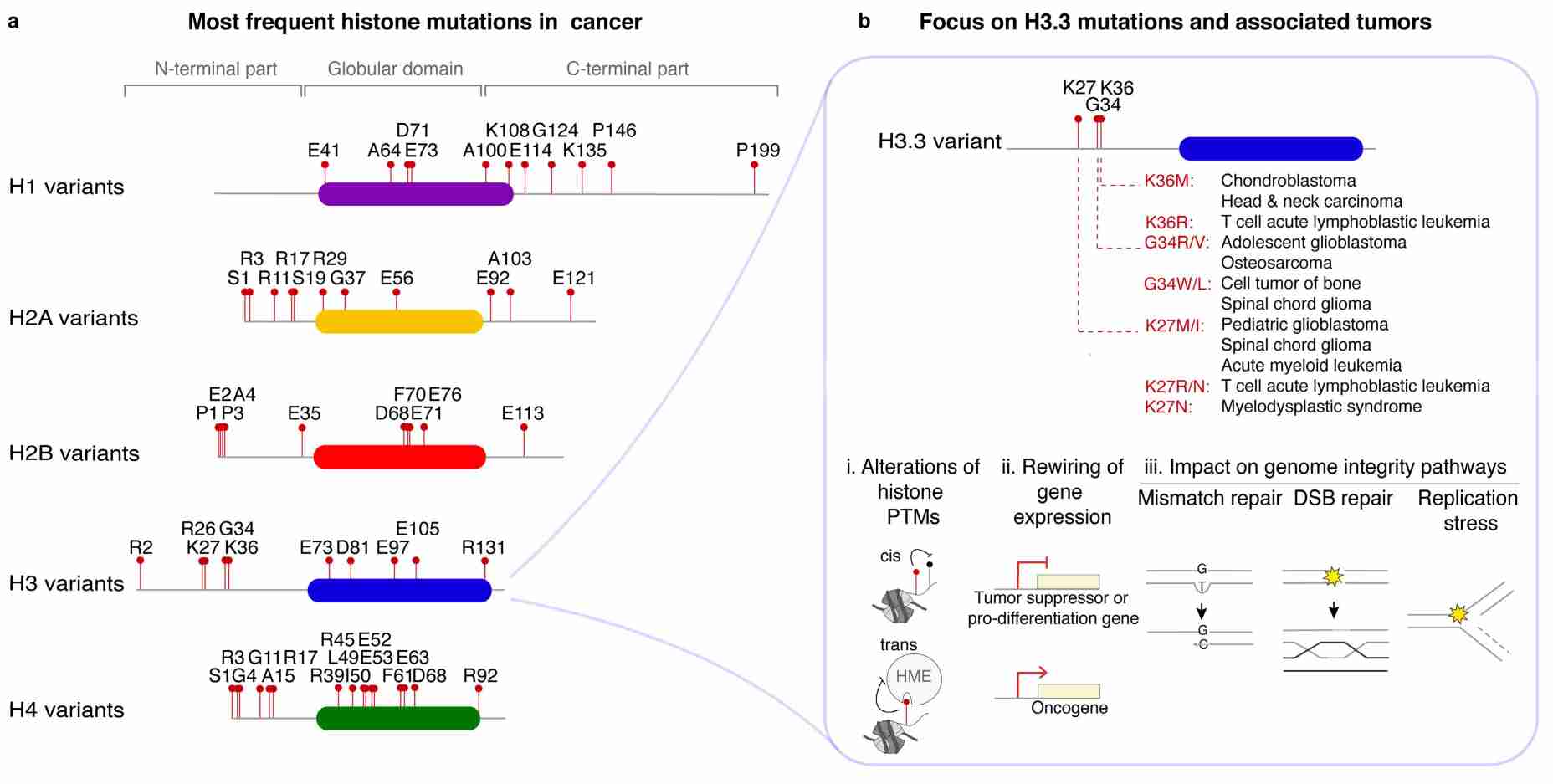 Fig.1 Histone variant point mutations in cancer. (Ferrand J, et al., 2020)
Fig.1 Histone variant point mutations in cancer. (Ferrand J, et al., 2020)Histone Modifications
Histone modifications refer to the addition or removal of chemical groups to histone proteins, which are key components of chromatin. Histone undergoes various post-translational modifications including acylation (e.g., acetylation, benzoylation, butyrylation, crotonylation, glutarylation, lactylation, and so on), ADP-ribosylation, dopaminylation, glycosylation, methylation, phosphorylation, serotonylation, sumoylation, and ubiquitination. Various histone-modifying enzymes have been shown to catalyze the addition or removal of a specific histone modification. Recently, histone tail cleavage by various enzymes has been regarded as an alternative modification to remove pre-existing tail modifications while retaining the histone folding region. Histone modifications exert critical effects on the interaction of different histones or histone–DNA interactions. Furthermore, histone modifications provide or remove binding sites for specific protein complexes, including histone-modifying enzymes, chromatin remodeling complexes, and transcription factors. Consequently, histone modifications regulate gene expression by altering the chromatin structure and protein–protein interaction.
Here are some of the common types of histone modifications:
| Modifications | Functions |
|---|---|
| Acetylation |
Acetylation involves the addition of an acetyl group to lysine residues (e.g., H3K9, H3K14, H3K18, H3K27, H4K5, H4K8, H4K12) on histone proteins. It is catalyzed by histone acetyltransferases (HATs) and can be reversed by histone deacetylases (HDACs). Acetylation neutralizes the positive charge of histones, thereby loosening their interaction with DNA. It generally correlates with gene activation and promotes an open chromatin structure, facilitating access to transcription factors and other regulatory proteins. Common sites of histone acetylation are K5, K9, and K13 on histone H2A; K5, K12, K15, and K20 on H2B; K9, K14, K18, K23, K27, K56, and K79 on H3; and K5, K8, K12, K16, and K91 on H4. The various acetylation sites of histones correspond to different functions. For example, H3K56 acetylation is involved in the regulation of nucleosome assembly, while H4K16 acetylation is involved in the regulation of nucleosome-mediated chromatin compaction, activation or inhibition of gene transcription, DNA damage repair, and other processes. |
| Methylation |
Methylation refers to the addition of methyl groups to histone lysine (e.g., H3K4, H3K9, H3K27) and arginine residues (e.g., H3R2, H3R17). It can be catalyzed by histone methyltransferases (HMTs) and removed by histone demethylases. Methylation can have different effects depending on the specific site and degree of methylation. Generally, methylation of H3K4 and H3K36 is associated with gene activation, whereas methylation of H3K9 and H3K27 is associated with gene repression. Methylation occurs mainly at K4, K9, K27, K36, and K79 on histone H3 and at K20 of H4. |
| Phosphorylation | Phosphorylation involves the addition of phosphate groups to serine, threonine, or tyrosine residues on histones (e.g., H3S10, H3T3). It is catalyzed by protein kinases and can be reversed by phosphatases. Phosphorylation of histones can impact chromatin structure and gene expression by influencing interactions with other proteins and promoting chromatin remodeling. |
| Ubiquitination |
Ubiquitination refers to the addition of ubiquitin, a small protein, to histone H2A and H2B lysine residues (e.g., H2AK119, H2BK120). It is catalyzed by ubiquitin ligases and can be reversed by deubiquitinases. Ubiquitination can have diverse effects depending on the specific residue and context. For example, the ubiquitination of H2BK120 is associated with gene activation, while the ubiquitination of H2AK119 is associated with gene repression. Most histones are monoubiquitinated rather than polyubiquitinated, which occurs mainly on H2A and H2B. |
| Sumoylation | Sumoylation involves the addition of small ubiquitin-like modifier (SUMO) proteins to lysine residues on histones (primarily H2BK126 and H3K9). It is catalyzed by SUMO ligases and can be reversed by SUMO proteases. Sumoylation can affect chromatin organization by altering protein-protein interactions and recruiting specific regulatory factors, affecting gene expression. |
Histones can experience various post-translational modifications in their protruding N-terminal tails but also within the C-terminal regions. These changes affect not only the overall compaction of chromatin but also gene expression.
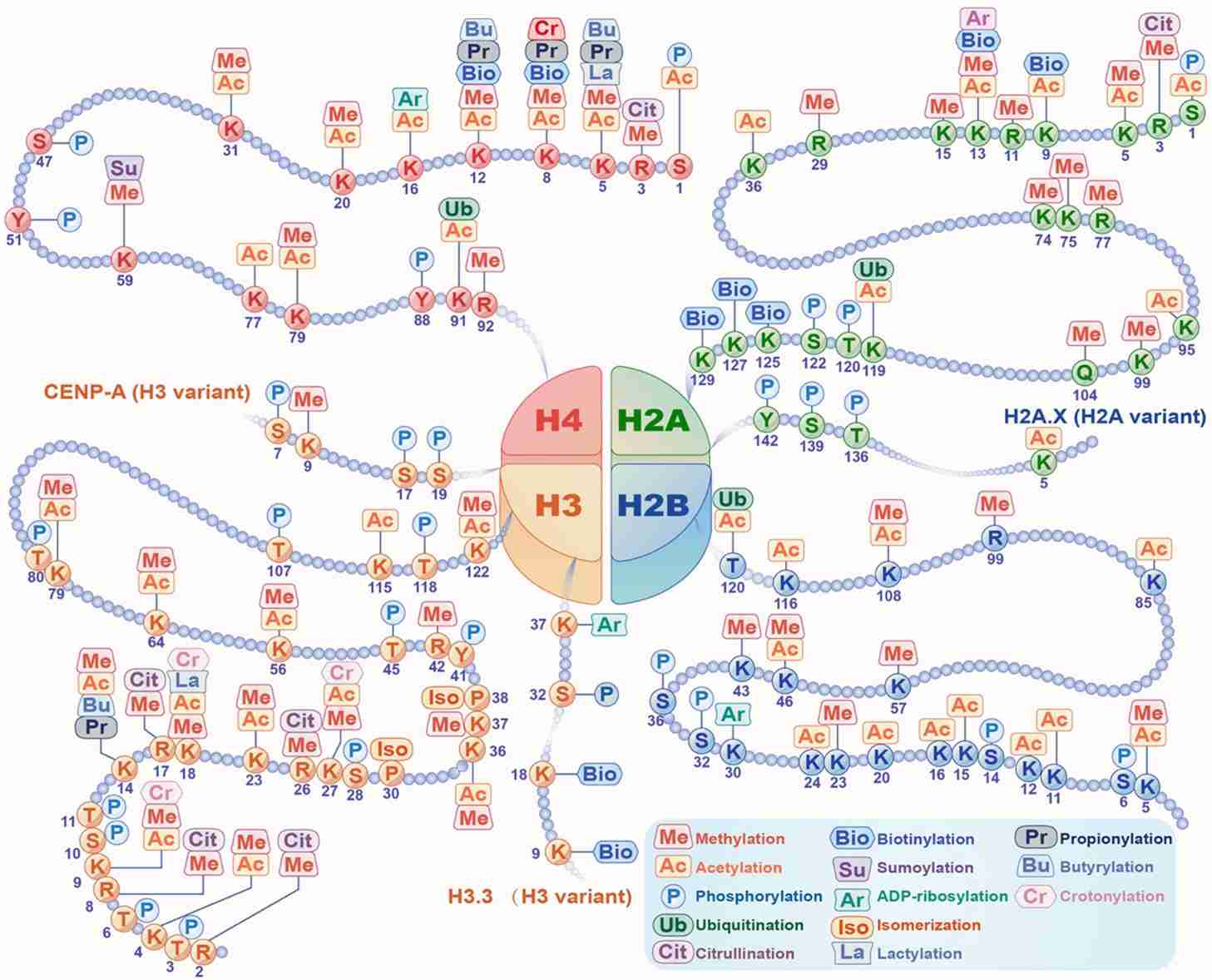 Fig.2 Histone modifications. (Yang Y, et al., 2022)
Fig.2 Histone modifications. (Yang Y, et al., 2022)Histone PTMs in Cancer Development and Neurological Disorders
Histones and their modifications have been extensively studied in the context of various diseases, and research in this area has provided valuable insights into disease development and potential therapeutic strategies. Here are some examples of the role of histones in disease and recent research progress:
| Histone PTMs | In cancer & neurological disorders |
|---|---|
| Histone Acetylation |
Acetylation of histone lysine residues by histone acetyltransferases (HATs) neutralizes the positive charge of histones, leading to an open chromatin structure and gene activation. In cancer, altered histone acetylation patterns can contribute to oncogenesis.
Altered histone acetylation patterns have been observed in several neurological disorders, influencing gene expression and neuronal function. Examples include:
|
| Histone Methylation |
Histone methylation, catalyzed by histone methyltransferases (HMTs), can have different effects depending on the specific lysine or arginine residue being modified and the degree of methylation. Examples include:
Abnormal histone methylation patterns have been linked to neurological disorders, affecting gene expression and synaptic plasticity. Examples include:
|
| Histone Phosphorylation |
Phosphorylation of histone residues can regulate gene expression and cellular processes. Aberrant histone phosphorylation has been implicated in cancer progression. For example:
Histone phosphorylation is involved in the regulation of neuronal activity and synaptic plasticity. Aberrant histone phosphorylation has been implicated in neurological disorders. Examples include:
|
| Histone Ubiquitination |
Ubiquitination of histones can modulate gene expression and chromatin structure. Altered histone ubiquitination has been linked to cancer development. For instance:
Histone ubiquitination is involved in the regulation of gene expression and synaptic function. Dysregulation of histone ubiquitination has been implicated in neurological disorders. Examples include:
|
| Histone Sumoylation |
Sumoylation, the attachment of small ubiquitin-like modifier (SUMO) proteins to histones, can regulate gene expression and chromatin structure. Dysregulation of histone sumoylation has been implicated in cancer. For example:
Histone sumoylation modulates gene expression and neuronal function. Abnormal histone sumoylation has been observed in neurological disorders. Examples include:
|
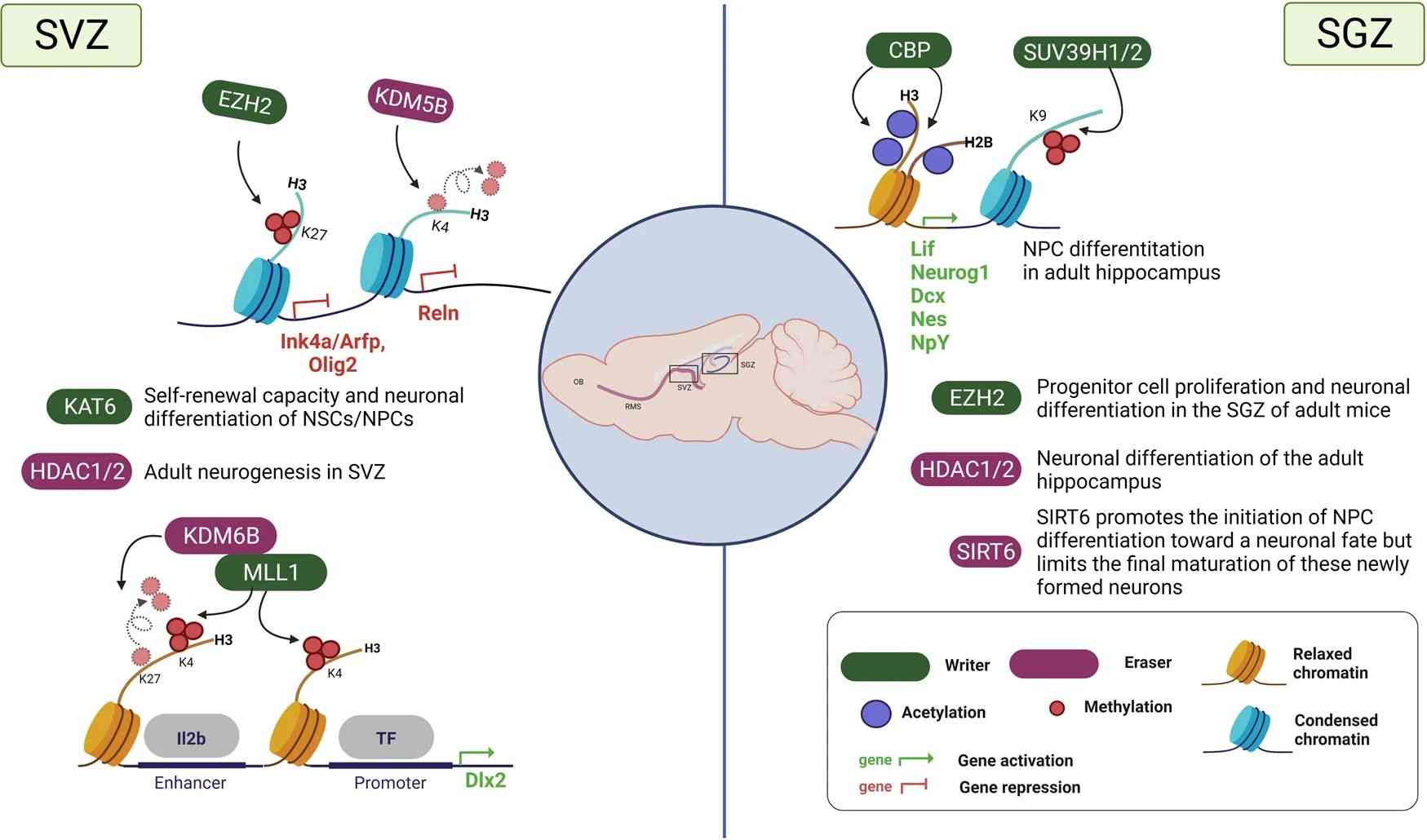 Fig.3 Histone modifications during adult neurogenesis. (Park J, et al., 2022)
Fig.3 Histone modifications during adult neurogenesis. (Park J, et al., 2022)Therapeutic Targeting of Histone PTMs
Therapeutic targeting of histone post-translational modifications (PTMs) has gained significant attention as a potential strategy for treating various diseases, including cancer and neurological disorders. Here are some approaches and examples of therapeutic targeting of histone PTMs:
Histone Deacetylase (HDAC) Inhibitors
HDAC inhibitors are compounds that block the activity of histone deacetylases, leading to increased histone acetylation and altered gene expression. HDAC inhibitors have shown promise in cancer treatment by promoting tumor cell differentiation, inducing cell cycle arrest, and enhancing immune responses. Examples of HDAC inhibitors include vorinostat, romidepsin, and panobinostat.
Histone Methyltransferase (HMT) Inhibitors
HMT inhibitors target enzymes responsible for histone methylation, leading to altered gene expression patterns. These inhibitors can be selective for specific methyltransferases or target a broader range of enzymes. HMT inhibitors have shown potential in cancer therapy by modulating gene expression and inhibiting tumor growth. Examples include EZH2 inhibitors (e.g., tazemetostat) targeting the histone methyltransferase EZH2 in certain cancers.
BET (Bromodomain and Extra-Terminal) Inhibitors
BET inhibitors selectively target bromodomain proteins that recognize acetylated histones, thereby affecting gene transcription. These inhibitors disrupt the interaction between bromodomain proteins and chromatin, leading to altered gene expression. BET inhibitors have shown promise in cancer treatment, particularly in hematological malignancies. Examples include JQ1 and OTX015.
Histone Demethylase Inhibitors
Histone demethylase inhibitors target enzymes involved in the removal of methyl groups from histones, thereby affecting gene expression. These inhibitors can selectively target specific demethylases or have a broader spectrum of activity. Histone demethylase inhibitors have shown potential in cancer therapy by modulating gene expression patterns. Examples include LSD1 inhibitors (e.g., GSK2879552) targeting the histone demethylase LSD1.
Targeting Reader Proteins
Reader proteins recognize and bind to specific histone PTMs, translating the epigenetic information into functional outcomes. Targeting reader proteins can disrupt the interaction between PTMs and their binding partners, leading to altered gene expression. Several small molecules and peptides have been developed to target reader proteins, and they hold therapeutic potential. Examples include inhibitors targeting bromodomain-containing proteins (e.g., BRD4 inhibitors) and methyl-lysine binding proteins (e.g., MBT domain inhibitors).
Combination Therapies
Combining therapies targeting different histone PTMs or combining epigenetic therapies with conventional treatments, such as chemotherapy or immunotherapy, can enhance therapeutic efficacy. Combinatorial approaches may provide synergistic effects and overcome resistance mechanisms.
It is important to note that the development and clinical use of histone PTM-targeting therapies are still in progress, and more research is needed to optimize their efficacy and minimize potential side effects. Additionally, the specific targeting strategies depend on the histone PTM involved, the disease context, and the availability of suitable inhibitors or modulators.
Case Study
Case 1: Kumar, A., Kumari, N., Sharma, U. et al. Reduction in H3K4me patterns due to aberrant expression of methyltransferases and demethylases in renal cell carcinoma: prognostic and therapeutic implications. Sci Rep 9, 8189 (2019).
Renal cell carcinoma (RCC) is a leading cause of cancer-related deaths among urological cancers. It is influenced by genetic and epigenetic factors. Histone methylations have been linked to renal tumorigenesis, but their clinical significance and underlying pathology are not well understood. In this study, researchers investigated the patterns of histone 3 lysine 4 (H3K4) methylation in clear cell RCC and its underlying pathology.
The authors attempted to establish any correlation between H3K4me1, H3K4me2, and H3K4me3 levels with metastasis of tumors. Remarkably, a reduction in all the three forms of H3K4me levels was observed with metastasis of the tumor. Further, the performance of H3K4me1, H3K4me2 and H3K4me3 in predicting metastasis was assessed using Receiver operative characteristics (ROC) curve, which showed that H3K4me3 might be a fair predictor for tumor metastasis.
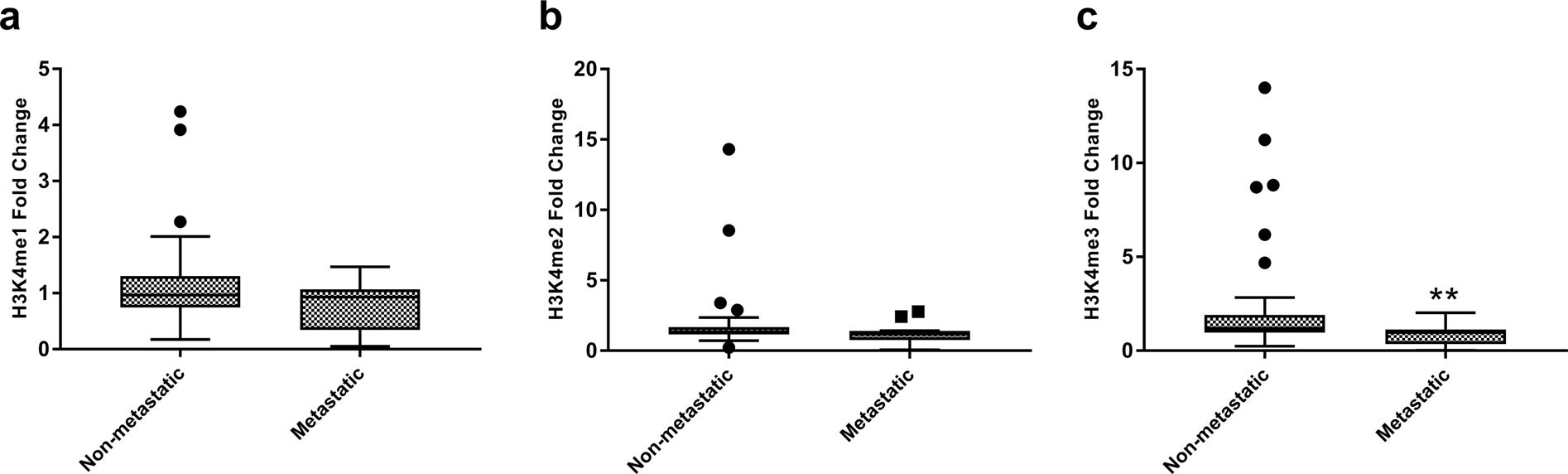 Fig.1 Global histone H3 lysine 4 methylation (H3K4me) levels between metastatic and primary tumors.
Fig.1 Global histone H3 lysine 4 methylation (H3K4me) levels between metastatic and primary tumors.Case 2: Hu S, Chapski DJ, Gehred ND, et al. Histone H1.0 couples cellular mechanical behaviors to chromatin structure. Nat Cardiovasc Res. 2024;3(4):441-459.
In this study, researchers investigated the distribution and role of linker histone H1.0 in the context of fibroblast activation. Unlike transcription factors or some modified core nucleosome histones, histone H1.0 is distributed fairly ubiquitously across the genome. The researchers performed chromatin immunoprecipitation sequencing (ChIP-seq) for histone H1.0 and found that it was depleted at genes that undergo altered expression after fibroblast activation, with a greater depletion observed in genes whose expression is increased. This suggests that the eviction of histone H1.0 is associated with chromatin relaxation.
The researchers confirmed the specific localization of histone H1.0 to several target genes of transforming growth factor beta (TGF-β) using chromatin immunoprecipitation followed by quantitative polymerase chain reaction (ChIP-qPCR). They also demonstrated that histone H1.0 plays a role in chromatin folding by performing nuclease digestion of genomic DNA. Overexpression of histone H1.0 increased the proportion of compacted DNA, while knockdown of histone H1.0 had the opposite effect. Interestingly, treatment with TGF-β caused the genome to adopt a more compact state, and knockdown of histone H1.0 reversed this effect.
Further experiments using targeted PCR for specific chromatin regions confirmed that the presence of histone H1.0 tends to compact those regions. Histone H1.0 overexpression or TGF-β treatment rendered these regions less accessible, while knockdown of histone H1.0 had the opposite effect.
These findings indicate that histone H1.0 establishes a microenvironment that influences the expression or repression of genes involved in stress-activated transcription. Perturbing the balance of histone H1.0 levels disrupts this microenvironment and prevents normal stress-activated transcription in fibroblast activation.
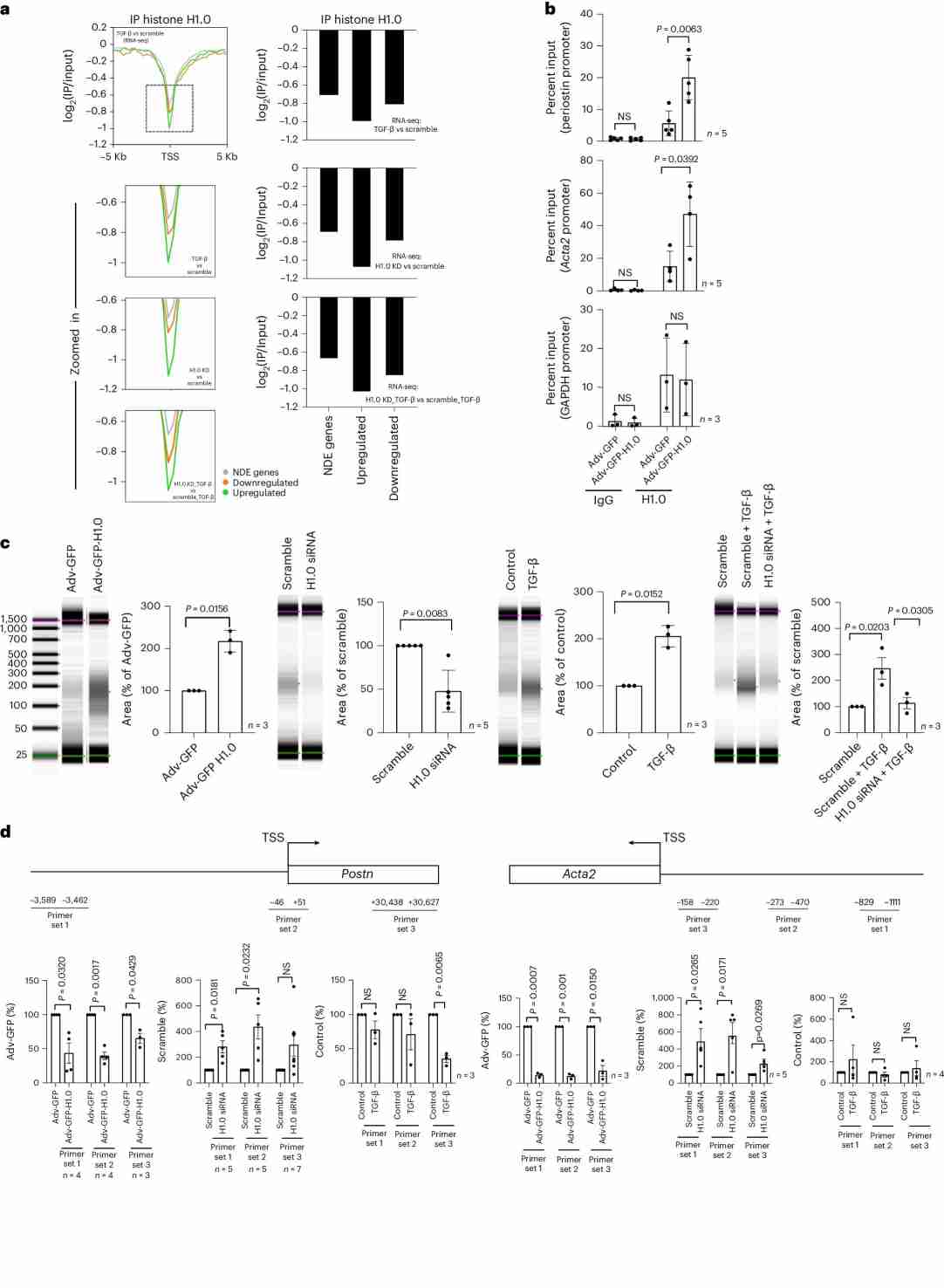 Fig.2 Histone H1.0 levels control chromatin fiber compaction.
Fig.2 Histone H1.0 levels control chromatin fiber compaction.Related References
- Zhang Y, Zhang Q, Zhang Y, Han J. The role of histone modification in DNA replication-coupled nucleosome assembly and cancer. International Journal of Molecular Sciences. 2023; 24(5):4939.
- Yang Y, Zhang M, Wang Y. The roles of histone modifications in tumorigenesis and associated inhibitors in cancer therapy. Journal of the National Cancer Center. 2022 Dec 1;2(4):277-90.
- Park J, Lee K, Kim K, et al. The role of histone modifications: from neurodevelopment to neurodiseases. Signal Transduction and Targeted Therapy, 2022, 7(1): 217.
- Ferrand J, Rondinelli B, Polo SE. Histone variants: guardians of genome integrity. Cells. 2020; 9(11):2424.
