B Cell Transcription Factors
Related Symbol Search List
Immunology Background
Background
B-cell transcription factors are a group of proteins that control the transcription of genes involved in B-cell development, signaling, and immune responses. They include both lineage-specific factors that are specific to B cells and factors shared with other cell types.
B-cell transcription factors orchestrate the stepwise development of B cells from hematopoietic stem cells in the bone marrow, as well as their maturation and activation in peripheral lymphoid tissues. They regulate the expression of genes involved in B-cell receptor (BCR) signaling, class switch recombination (CSR), somatic hypermutation (SHM), antibody production, and immune response modulation.
B-cell transcription factors can be broadly categorized into lineage-specifying factors and factors involved in B-cell differentiation and function.
- Lineage-specifying factors: These are critical for the commitment of multipotent progenitor cells to the B-cell lineage. Examples include E2A (E12/E47), EBF1 (Early B-cell Factor 1), Pax5 (Paired box protein 5).
- Factors involved in B-cell differentiation and function: These factors regulate various aspects of B-cell development, activation, and immune responses. Examples include Ikaros family proteins (Ikaros, Aiolos), BCL6 (B-cell lymphoma 6), IRF4 (Interferon Regulatory Factor 4), Blimp-1 (PRDM1).
- B-cell transcription factors regulate gene expression by interacting with DNA sequences, recruiting co-activators or co-repressors, and modifying chromatin structure. They control the expression of key genes involved in B-cell development, survival, proliferation, differentiation, and immune responses. By coordinating the activity of these transcription factors, B cells can undergo lineage commitment, antibody diversification, and generate appropriate immune responses to antigens.
Understanding the functions and interactions of B-cell transcription factors is crucial for unraveling the regulatory networks that govern B-cell development, differentiation, and immune responses. Dysregulation of these factors can lead to immune disorders, lymphomas, and autoimmune diseases. Therefore, studying B-cell transcription factors is important for both basic research and the development of targeted therapies for B-cell-related disorders.
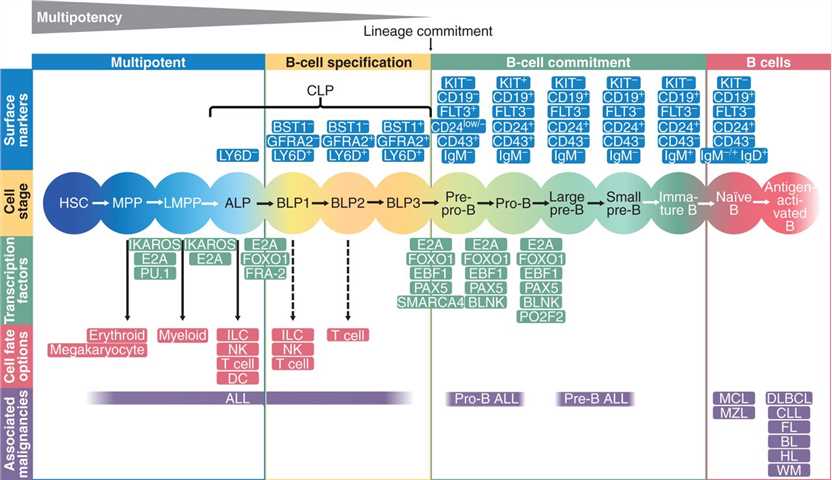 Fig.1 Transcriptional regulation determines normal B-cell development. (Fischer U, et al., 2020)
Fig.1 Transcriptional regulation determines normal B-cell development. (Fischer U, et al., 2020)Key B Cell Transcription Factors
| Types | Functions |
|---|---|
| Lineage-Specifying Factors |
|
| Transcription Factors Involved in B-Cell Differentiation and Function |
|
These transcription factors collectively modulate gene expression programs necessary for B-cell lineage commitment, development, differentiation, and immune responses. Their precise functions and interactions ensure the proper functioning of B cells in the immune system.
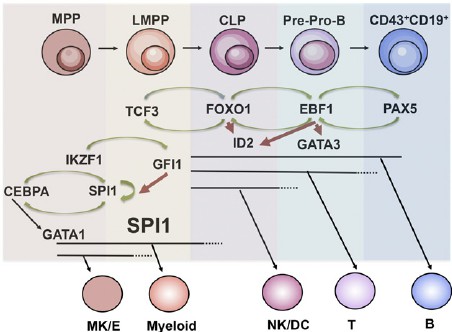 Fig.2 Regulatory networks in early B-cell development. Schematic drawing over regulatory networks involved in B-cell priming, specification, and commitment. (Somasundaram R, et al., 2015)
Fig.2 Regulatory networks in early B-cell development. Schematic drawing over regulatory networks involved in B-cell priming, specification, and commitment. (Somasundaram R, et al., 2015)Regulatory Mechanisms and Signaling Pathways of B Cell Transcription Factors
The regulatory mechanisms and signaling pathways of B-cell transcription factors involve a complex interplay of various factors and pathways. Here are some key regulatory mechanisms and signaling pathways that influence B-cell transcription factors:
| Regulatory mechanisms & signaling pathways | Details |
|---|---|
| B-Cell Receptor (BCR) Signaling | BCR signaling is a crucial pathway that regulates B-cell transcription factors. Upon antigen recognition by the BCR, a cascade of signaling events is initiated, leading to the activation of transcription factors. The key signaling molecules involved in BCR signaling include Syk, Bruton's tyrosine kinase (BTK), phospholipase C gamma 2 (PLCγ2), and protein kinase C (PKC). Activation of BCR signaling pathways ultimately leads to the activation or modulation of various B-cell transcription factors, including NF-κB, AP-1 (activator protein 1), and IRF4. |
| Cytokine Signaling | Cytokines play a crucial role in modulating B-cell transcription factors. Cytokines such as IL-4, IL-6, and IL-21 influence B-cell development, differentiation, and function by activating specific signaling pathways. These pathways can activate or regulate transcription factors such as STAT (signal transducer and activator of transcription) proteins, which then modulate gene expression programs in B cells. |
| Co-receptors and Co-stimulatory Molecules | Co-receptors and co-stimulatory molecules expressed on B cells, such as CD19, CD40, and CD80/86 (B7-1/B7-2), provide additional signals that regulate B-cell transcription factors. For example, CD40 signaling, initiated by interaction with CD40 ligand (CD40L) on T cells, activates NF-κB and AP-1, which are important transcription factors for B-cell activation, proliferation, and differentiation. |
| PI3K-Akt-mTOR Pathway | The phosphoinositide 3-kinase (PI3K)-Akt-mTOR pathway is involved in regulating B-cell transcription factors in response to various stimuli. Activation of this pathway influences B-cell survival, proliferation, and metabolism. Akt, a downstream effector of PI3K, can phosphorylate and modulate the activity of transcription factors such as FOXO (forkhead box O) proteins, which play a role in B-cell development and apoptosis. |
| Epigenetic Regulation | Epigenetic modifications, including DNA methylation, histone modifications, and chromatin remodeling, are essential for regulating B-cell transcription factors. These modifications can affect the accessibility of transcription factor binding sites on DNA, thereby influencing gene expression. For example, DNA methylation can regulate the expression of transcription factors involved in B-cell development and differentiation. |
| Feedback Regulation | B-cell transcription factors often engage in feedback loops to regulate their own expression or that of other transcription factors. For instance, PAX5 regulates its own expression and is involved in a network of transcription factors, including EBF1, E2A, and IRF4, which collectively regulate B-cell development and function. |
| Microenvironmental Signals | The B-cell microenvironment, such as interactions with stromal cells, follicular dendritic cells, and other immune cells, provides important signals that influence B-cell transcription factors. These signals can come from chemokines, adhesion molecules, and other factors present in the microenvironment, which can activate signaling pathways and modulate transcription factor activity. |
These are just a few examples of the regulatory mechanisms and signaling pathways involved in the regulation of B-cell transcription factors. The intricate interplay between these factors and pathways ensures proper B-cell development, differentiation, and immune response, allowing B cells to fulfill their roles in humoral immunity.
Association Between B-Cell Transcription Factors and Disease
B-Cell Acute Lymphoblastic Leukemia (B-ALL)
PAX5 is a critical transcription factor involved in B-cell development and maintenance of B-cell identity. Mutations or deletions in the PAX5 gene are frequently observed in B-ALL. Loss of PAX5 function disrupts normal B-cell differentiation and leads to the accumulation of immature B cells, contributing to leukemogenesis.
Diffuse Large B-Cell Lymphoma (DLBCL) and Burkitt Lymphoma
FOXP1 is a transcription factor that regulates B-cell development and function. Aberrant expression or mutations in FOXP1 have been associated with DLBCL and Burkitt lymphoma. Dysregulation of FOXP1 can affect cell cycle control, apoptosis, and DNA repair pathways, contributing to lymphomagenesis and influencing the aggressive behavior of these B-cell lymphomas.
Multiple Myeloma (MM)
PRDM1, also known as Blimp-1, is a transcription factor essential for plasma cell differentiation. Dysregulation of PRDM1 has been implicated in multiple myeloma. Reduced expression or loss of PRDM1 can lead to impaired plasma cell differentiation and the accumulation of malignant plasma cells, contributing to disease progression.
Hyper-IgM Syndrome Type 2 (HIGM2)
Activation-induced cytidine deaminase (AICDA) is a transcription factor involved in somatic hypermutation (SHM) and class switch recombination (CSR) processes in B cells. Defects in the AICDA gene result in HIGM2, a rare immunodeficiency disorder characterized by defective antibody production. Impaired AICDA function prevents the generation of high-affinity antibodies and compromises the immune response.
Hematological Malignancies (including Acute Myeloid Leukemia and B-cell lymphomas)
SPI1 is a transcription factor critical for normal hematopoiesis, including B-cell development. Dysregulation of SPI1 has been associated with various hematological malignancies, including acute myeloid leukemia (AML) and B-cell lymphomas. Altered SPI1 expression or mutations can disrupt normal B-cell differentiation and contribute to leukemogenesis.
B-Cell Precursor Acute Lymphoblastic Leukemia (BCP-ALL)
Early B-cell factor 1 (EBF1) and EBF2 are transcription factors crucial for B-cell lineage commitment and development. Disruptions in EBF1 or EBF2 function have been observed in BCP-ALL, a subtype of ALL. Loss of EBF1 or EBF2 can interfere with normal B-cell differentiation and contribute to the development of BCP-ALL.
These examples highlight the specific roles of B-cell transcription factors in various diseases. Dysregulation or mutations in these transcription factors can disrupt normal B-cell development, differentiation, and function, leading to the development or progression of specific diseases. It's important to note that these associations are not exhaustive, and the involvement of transcription factors can vary across different subtypes or variations of the diseases mentioned.
Case Study
Case 1: Pang SH, Minnich M, Gangatirkar P, et al. PU.1 cooperates with IRF4 and IRF8 to suppress pre-B-cell leukemia. Leukemia. 2016;30(6):1375-1387.
The Ets family transcription factor PU.1 and the interferon regulatory factor (IRF)4 and IRF8 regulate gene expression by binding tocomposite DNA sequences known as Ets/interferon consensus elements. Although all three factors are expressed from the onset ofB-cell development, single deficiency of these factors in B-cell progenitors only mildly impacts on bone marrow B lymphopoiesis.
Here the authors test if PU.1 also cooperates with IRF4 during B-cell develop-ment PU.1fl/flMb1cremice were crossed to Irf4−/−mice to generatePU.1/IRF4 DKO mice, which lack both proteins only in the B-cellcompartment. Similar to IRF8-deficient mice, IRF4 loss resulted in amoderate increase in pro-/pre-B cells and a twofold decrease inrecirculating B cells. Like PU.1/IRF8 deficiency, a severereduction of recirculating B cells was observed in PU.1/IRF4 DKOmice (b, g). Analysis of Irf4−/−mice lacking asingle copy of PU.1 (PU.1fl/+Mb1creIrf4−/−) demonstrated a dosedependency of this Ets-IRF complex as the loss of transitionaland recirculating B cells was more pronounced than in Irf4−/−mice (f, g). Taken together, these results identifyan important collaboration between PU.1 and IRF4 or IRF8 duringBM B-cell development and maturation.
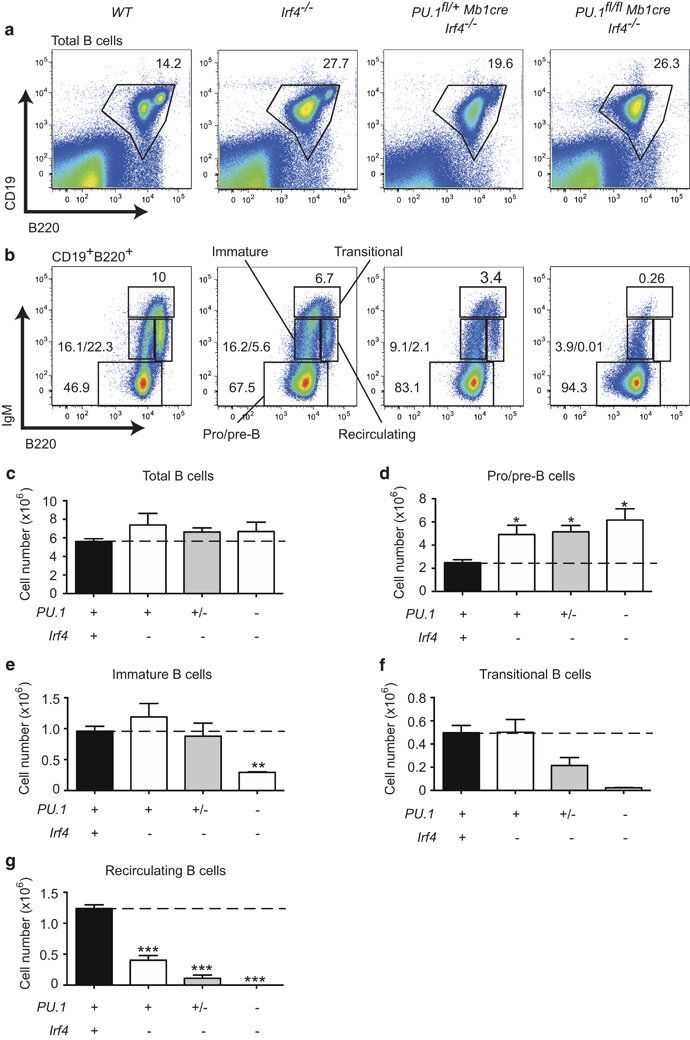 Fig.1 Analysis of B-cell development in the absence of PU.1 and IRF4.
Fig.1 Analysis of B-cell development in the absence of PU.1 and IRF4.Case 2: Xia Y, Xu-Monette ZY, Tzankov A, et al. Loss of PRDM1/BLIMP-1 function contributes to poor prognosis of activated B-cell-like diffuse large B-cell lymphoma. Leukemia. 2017;31(3):625-636.
PRDM1 /BLIMP-1, a master regulator of plasma-cell differentiation, is frequently inactivated in activated B-cell-like (ABC) diffuse large B-cell lymphoma (DLBCL) patients. The authors assessed a large cohort of patients with de novo DLBCL for PRDM1/BLIMP-1 deletion, mutation, and protein expression. And assessed correlation of PRDM1 mutations with poor prognosis for DLBCL. (a) In ABC-DLBCL, patients with PRDM1 mutations had shorter OS and PFS durations than did patients with wild-type PRDM1. (b) PRDM1 mutations within exons 1 and 2 had even greater poor impact on OS and PFS compared to did the overall mutation cohort. (c) The detailed distribution of PRDM1 mutations within exons 1 and 2. The authors found that patients with exons 1 and 2 PRDM1 mutations were more likely to have overexpression of both Myc and Bcl-2 than were patients with exons 3–7 mutations in the overall cohort and those with wild-type PRDM1 in ABC-DLBCL but not in those with GCB-DLBCL. Multivariate analysis of clinical parameters, Myc/Bcl-2 expression scores, and PRDM1 mutations indicated that Myc/Bcl-2 co-expression, but not PRDM1 mutations, was an independent prognostic factor.
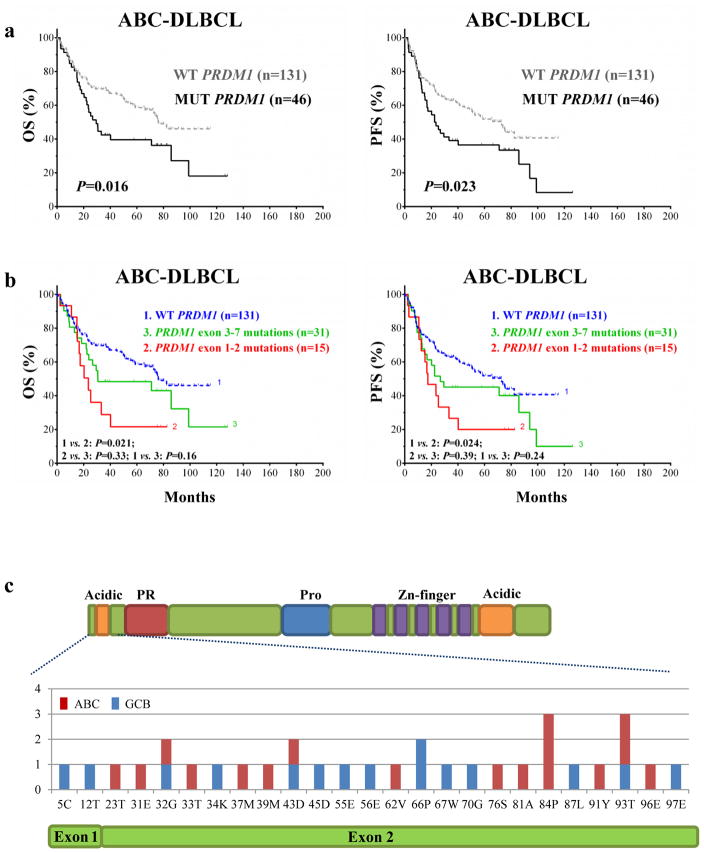 Fig.2 Correlation of PRDM1 mutations with poor prognosis for DLBCL.
Fig.2 Correlation of PRDM1 mutations with poor prognosis for DLBCL.References
- Fischer U, Yang JJ, Ikawa T, et al. Cell Fate Decisions: The Role of Transcription Factors in Early B-cell Development and Leukemia. Blood Cancer Discov. 2020;1(3):224-233.
- Somasundaram R, Prasad MA, Ungerbäck J, Sigvardsson M. Transcription factor networks in B-cell differentiation link development to acute lymphoid leukemia. Blood. 2015;126(2):144-152.
