SIRT2
-
Official Full Name
sirtuin 2 -
Overview
This gene encodes a member of the sirtuin family of proteins, homologs to the yeast Sir2 protein. Members of the sirtuin family are characterized by a sirtuin core domain and grouped into four classes. The functions of human sirtuins have not yet been determined; however, yeast sirtuin proteins are known to regulate epigenetic gene silencing and suppress recombination of rDNA. Studies suggest that the human sirtuins may function as intracellular regulatory proteins with mono-ADP-ribosyltransferase activity. The protein encoded by this gene is included in class I of the sirtuin family. Several transcript variants are resulted from alternative splicing of this gene. [provided by RefSeq, Jul 2010] -
Synonyms
SIRT2;sirtuin 2;SIR2;SIR2L;SIR2L2;NAD-dependent protein deacetylase sirtuin-2;sirtuin-2;sirtuin type 2;SIR2-like protein 2;sir2-related protein type 2;silent information regulator 2;regulatory protein SIR2 homolog 2;NAD-dependent deacetylase sirtuin-2
Recombinant Proteins
- Human
- Mouse
- Chicken
- Rat
- Zebrafish
- E.coli
- Mammalian Cell
- Insect Cell
- Sf9 Insect Cell
- HEK293
- His
- His&T7
- Non
- HIS
- GST
- His&Fc&Avi
Background
What is SIRT2 Protein?
SIRT2 is a human enzyme that relies on NAD+ to do its thing, acting as both a lysine deacetylase and defatty-acylase. Essentially, it helps take off specific chemical groups from proteins. What's interesting about SIRT2 is that it tends to form pairs, or dimers, both in test tubes and in living cells. This pairing seems to change how well it can handle different types of protein modifications. The process was studied using various lab techniques like size exclusion chromatography and even X-ray crystal structures. They found that when SIRT2 pairs, it can split into single units if it encounters long fatty acid chains, but not with shorter ones like acetyl-lysine. They also discovered a mutant version of SIRT2 that has a harder time pairing up, which messes with its usual activity. Interestingly, certain inhibitors like ascorbyl palmitate can break apart these pairs, affecting how SIRT2 does its job. So, SIRT2's ability to pair up is a big part of how it functions, and this can be tweaked either through genetic changes or by using specific inhibitors.
What is the Function of SIRT2 Protein?
SIRT2 is a protein that acts as an NAD±dependent enzyme, known for removing acyl groups from proteins, like a deacetylase and defatty-acylase. It's known to pair up, or dimerize, both in solutions and inside cells. This dimerization is crucial because it influences how SIRT2 interacts with various acyl-modified substrates. For instance, its structure can shift from dimers to single units when handling long fatty acylated substrates, though this isn't the case with acetyl-lysine. Interestingly, a specific mutant version of SIRT2 that struggles to dimerize showed typical activity in breaking down fatty acyl groups but was less effective at deacetylation. This suggests that forming dimers is key for SIRT2's deacetylase function. Additionally, ascorbyl palmitate, an inhibitor, can split these dimers, pointing to a new method of controlling SIRT2's activity that can be tweaked either through genetic changes or drugs.

Fig1. SIRT2 participates in tumor inhibition and promotion. (Yan Wang, 2019)
SIRT2 Related Signaling Pathway
SIRT2 is a protein that primarily acts like a switchboard for managing cellular processes, relying on NAD+ to function. It's usually hanging out in the cytoplasm, where it helps control structures like microtubules, but it can pop into the nucleus when needed. While there, it plays a role in organizing DNA structures and responding to DNA damage. This enzyme's activity is tied to multiple signaling pathways, including ones involved in metabolism, inflammation, cell cycle regulation, and stress responses. For instance, SIRT2 can influence insulin sensitivity by acting on certain proteins, and its inhibition has shown promise in models of neurodegenerative diseases, suggesting it might have protective roles under specific conditions. It's also linked to pathways that either support or suppress tumor growth, showcasing its dual roles based on the cellular environment.
SIRT2 Related Diseases
SIRT2 is a protein that's been spotlighted in various diseases, largely because it plays a part in how cells age and die, tweaks metabolism, and handles stress responses. In neurodegenerative disorders like Parkinson's and Huntington's, SIRT2's involvement isn't entirely clear-cut, but it's thought to mess with nerve function and survival, potentially worsening these conditions. As for metabolic issues, SIRT2 seems to help control fat creation and insulin sensitivity, hinting it might act as a shield against diabetes. On the cancer front, SIRT2 could be a double-edged sword; some say it slows down tumors, while others think it might do the opposite, possibly letting cancer cells dodge death. So, SIRT2's role in diseases is quite a juggling act and is still under the microscope.
Bioapplications of SIRT2
SIRT2, a real multitasker in the lab and industry, is finding its way into varied applications. As a recombinant protein, it's being harnessed in research settings to explore its role in cellular processes, shedding light on its potential impact on diseases. Scientists also tap into its capabilities for innovative industrial production techniques. By tweaking SIRT2's functions, both researchers and industry experts aim to unlock new possibilities, making it a key player in both scientific discoveries and practical applications.
Case Study
Case Study 1: Jie Yang, 2023
Human SIRT2, an enzyme relying on NAD+, acts on lysines by removing acetyl and fatty acyl groups. We've found SIRT2 tends to form dimers both in solutions and cells, influencing how it interacts with different substrates. Techniques like size exclusion chromatography showed this dimerization. Interestingly, these dimers break into monomers with long fatty acyl substrates but stay intact with acetyl-lysine. By examining crystal structures, we identified a SIRT2 double mutant (Q142A/E340A) that struggles to dimerize, confirmed using chemical techniques. This mutant maintains its defatty-acylase activity but loses some deacetylase ability, emphasizing that dimerization is crucial for SIRT2's deacetylase function. We also discovered that SIRT2 dimers can be disrupted by the inhibitor ascorbyl palmitate. Our research reveals that SIRT2's oligomeric state and substrate preference are novel regulatory mechanisms, which can be influenced genetically or pharmacologically.
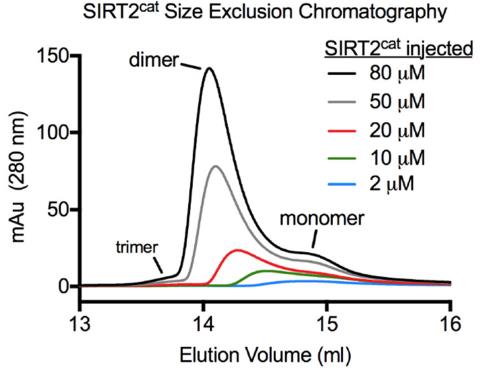
Fig1. Analytical SEC chromatograms were collected after injecting 100 μL of the indicated SIRT2cat concentrations.
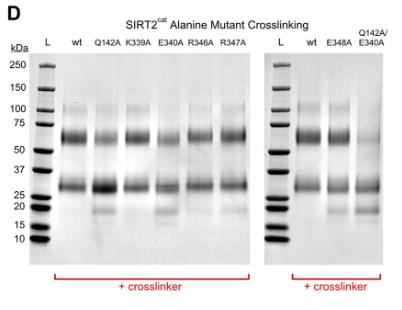
Fig2. Cross-linking experiment with Bis(NHS)-PEG5 and 10 μM SIRT2cat.
Case Study 2: Matthew J. G. Eldridge, 2020
Sirtuin-2 (SIRT2) is a protein that gets involved in a bunch of cell stuff like the cell cycle, metabolism, cancer, and bacterial infections. It usually hangs out in the cytoplasm but can also do its thing in the nucleus. Its location and interactions are key to understanding its functions. This study used mass spectrometry to explore SIRT2's interactions across whole cells and specific areas like the cytoplasm, nucleus, and chromatin. We discovered new partners for SIRT2, including several proteins involved in nuclear import. It turns out that various importins, including IPO7, interact with SIRT2, aiding its movement into the nucleus, which is crucial for SIRT2's role in deacetylating H3K18 during bacterial infections. Additionally, SIRT2's unstructured C-terminus seems to negatively influence its binding with importins and its nuclear transport.
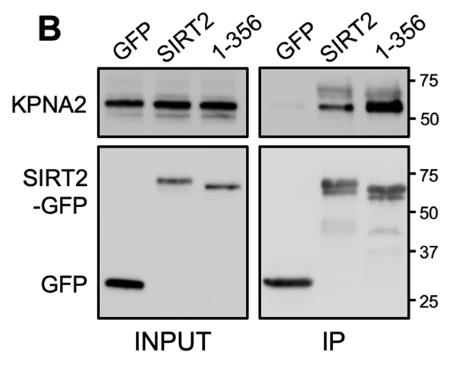
Fig3. Cell lysates (INPUT) and IP fractions were immunoblotted for with antibodies against GFP.
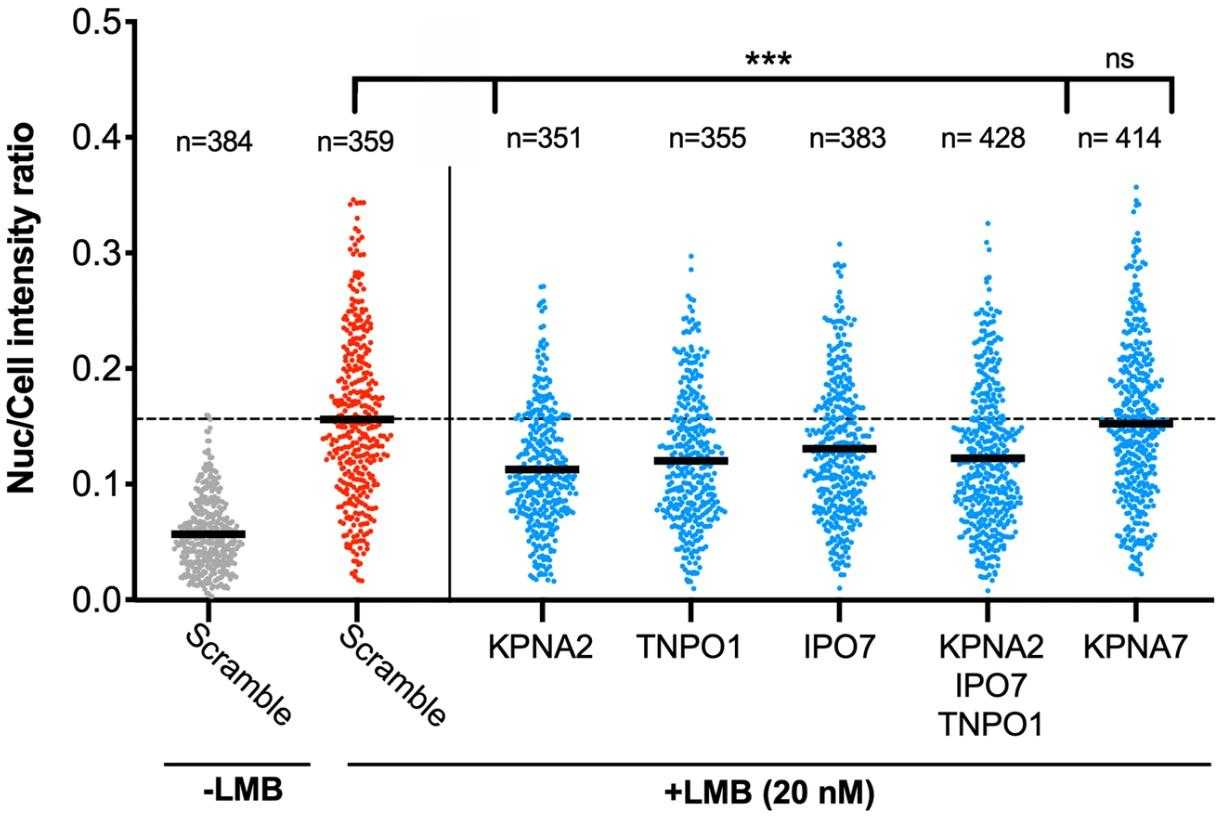
Fig4. Graphs show nuclear: whole cell intensity ratio of SIRT2-GFP.
Quality Guarantee
High Purity
.jpg)
Fig1. SDS-PAGE (SIRT2-31405TH)
.
.jpg)
Fig2. SDS-PAGE (SIRT2-647H)
Involved Pathway
SIRT2 involved in several pathways and played different roles in them. We selected most pathways SIRT2 participated on our site, such as Signaling events mediated by HDAC Class I,Signaling events mediated by HDAC Class III, which may be useful for your reference. Also, other proteins which involved in the same pathway with SIRT2 were listed below. Creative BioMart supplied nearly all the proteins listed, you can search them on our site.
| Pathway Name | Pathway Related Protein |
|---|---|
| Signaling events mediated by HDAC Class I | GATA2,TFCP2,MXD1,SIRT7,FKBP3,NR2C1,WDR77,GATA1,SIRT5,PRMT5 |
| Signaling events mediated by HDAC Class III | SIRT7 |
Protein Function
SIRT2 has several biochemical functions, for example, NOT NAD+ ADP-ribosyltransferase activity,NAD+ binding,NAD-dependent histone deacetylase activity. Some of the functions are cooperated with other proteins, some of the functions could acted by SIRT2 itself. We selected most functions SIRT2 had, and list some proteins which have the same functions with SIRT2. You can find most of the proteins on our site.
| Function | Related Protein |
|---|---|
| protein binding | COX4I2,XBP1,DPYD,CHMP4A,FGFRL1B,KRTAP5-9,PPIL1,P2RY6,ITGB2,IRAK1BP1 |
| beta-tubulin binding | PDCD5,GABARAPL2,B4GALT1,RANBP10,PIFO,GABARAP,BBS4,RACGAP1,CCT5,FGF13 |
| NAD+ binding | SIRT6,GLUD1,SIRT1,UXS1,HPGD,SIRT7,SIRT3,HADH,SIRT4,SIRT5 |
| NAD-dependent histone deacetylase activity | SIRT1,SIRT6 |
| NOT NAD+ ADP-ribosyltransferase activity | SIRT3,SIRT5,SIRT1 |
| histone deacetylase binding | SKOR2,PKN2,RUNX2,PRDM1,NUDT21,NCOR2,USF1,HDAC5,SP2,HDAC3 |
| protein deacetylase activity | HDAC10,HDAC2,SIRT1,HDAC9,HDAC4,HDAC1,HDAC5,HDAC3,SIN3A |
| tubulin deacetylase activity | HDAC6 |
| zinc ion binding | DTX1,TRIM4,MMP11,ADAM19,DIDO1,UQCRC2A,SKI,MICAL1,TRIM28,MORC1 |
Interacting Protein
SIRT2 has direct interactions with proteins and molecules. Those interactions were detected by several methods such as yeast two hybrid, co-IP, pull-down and so on. We selected proteins and molecules interacted with SIRT2 here. Most of them are supplied by our site. Hope this information will be useful for your research of SIRT2.
BUB1B;KAT2B;RELA;CDC14B;G6PD;RIPK3;MYOD1;HDAC6
Resources
Research Area
Histone Deacetylases (HDACs) and Associated ProteinsAlzheimer's Disease
Growth Hormone/IGF-I Axis
Cancer Stem Cell Transcription Factors
Other Autophagy-related Proteins
Related Services
Related Products
References
- Yeung, F; Ramsey, CS; et al. Regulation of the mitogen-activated protein kinase kinase (MEK)-1 by NAD(+)-dependent deacetylases. ONCOGENE 34:798-804(2015).
- Bajpe, PK; Prahallad, A; et al. A chromatin modifier genetic screen identifies SIRT2 as a modulator of response to targeted therapies through the regulation of MEK kinase activity. ONCOGENE 34:531-536(2015).


