SWI/SNF Family
Related Symbol Search List
Immunology Background
Background
In the field of molecular biology, the SWI/SNF (SWItch/Sucrose Non-Fermentable) subfamily represents a set of ATP-dependent chromatin remodeling complexes present in eukaryotic organisms. Essentially, these complexes consist of a collection of proteins that collaborate to restructure how DNA is packaged. The assemblage comprises various proteins stemming from the SWI and SNF genes (SWI1, SWI2/SNF2, SWI3, SWI5, SWI6), along with additional polypeptides. One of its key features is the possession of a DNA-activated ATPase activity capable of disrupting histone-DNA interactions within reconstituted nucleosomes in an ATP-reliant manner, although the specific mechanisms underlying this structural modification remain unclear.
The SWI/SNF subfamily plays a pivotal role in rearranging nucleosomes, leading to their expulsion and/or movement. This repositioning of nucleosomes facilitates easier access to chromatin, allowing specific transcription factors to bind effectively. Consequently, this process enables the activation or repression of genes.
SWI/SNF analogs in humans are known as BRG1 or BRM-associated factor, abbreviated BAF (SWI/SNF-A), and Polybromo-associated BAF, also known as PBAF (SWI/SNF-B). Proteins similar to SWI/SNF exist in Drosophila and are referred to as Brahma-associated protein (BAP) and Polybromo-associated BAP (PBAP). Recently, a new type of SWI/SNF complex, known as the GBAF or non-canonical BAF complex, has been discovered by Alpsoy et al. and Mashtalir et al. This complex does not contain ARID1A/B and ARID2, but instead includes unique subunits BRD9 and either GLTSCR1 or GLTSCR1L. All SWI/SNF complexes feature a catalytic ATPase subunit (SMARCA4/BRG1 or SMARCA2/BRM) and core subunits (SMARCC and SMARCD proteins). The assembly of these complexes is a highly ordered process, with the core module serving as a platform for recruiting complex-specific subunits and the ATPase module capping off the assembly.
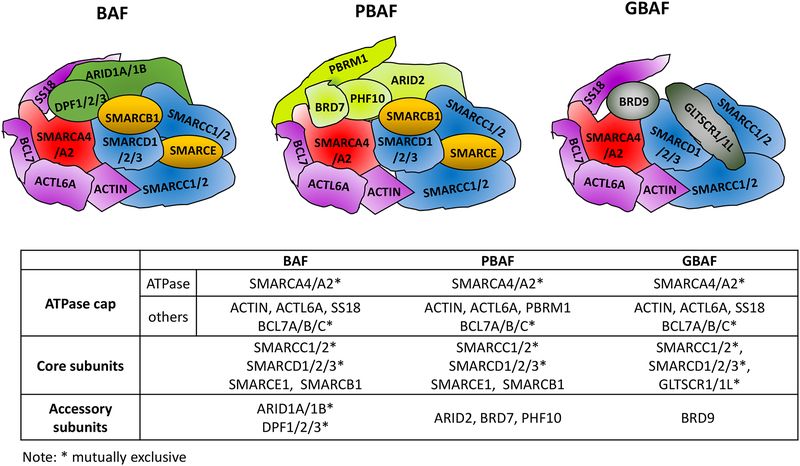 Fig.1 Scheme of three classes of SWI/SNF complex. (Wang Y, et al., 2020)
Fig.1 Scheme of three classes of SWI/SNF complex. (Wang Y, et al., 2020)Key Structural Features of SWI/SNF Family
The SWI/SNF family of chromatin remodeling complexes is characterized by a modular structure comprising several subunits with distinct functions and domains. These subunits interact to remodel chromatin structure, regulate gene expression, and participate in various cellular processes. Here is an overview of the structural characterization of the SWI/SNF family:
- The SWI/SNF family exhibits a modular architecture with specific subunits performing distinct functions within the complex.
- Interactions between subunits and their domains enable the complex to bind DNA, remodel chromatin, and regulate gene expression.
- The structural diversity within the SWI/SNF family allows for flexibility in responding to different cellular contexts and modulating gene expression patterns in a dynamic manner.
| Subunits | Structural features |
|---|---|
| ATPase Subunits |
|
| Core Subunits |
|
| Regulatory Subunits |
|
| DNA-Binding Subunits |
|
| ARID Domain-Containing Subunits |
|
| Bromodomain-Containing Subunits |
|
| Accessory Subunits |
|
Understanding the structural organization of the SWI/SNF family provides insights into how these complexes function in chromatin remodeling, gene regulation, and cellular processes crucial for development, differentiation, and response to environmental cues.
Functions of SWI/SNF Family
The SWI/SNF complexes plays a pivotal role in various cell biology and biochemical processes, including transcriptional regulation, chromosomal remodeling, and epigenetics. Here is a detailed examination of how the SWI/SNF family influences these critical cellular processes:
| Functions | Details |
|---|---|
| Transcriptional Regulation |
Facilitating Transcription Factor Access: SWI/SNF complexes remodel chromatin structure to make DNA regions accessible to transcription factors. This process allows regulatory proteins to bind to specific gene promoters or enhancers, thereby activating or repressing gene transcription. Enhancing RNA Polymerase Activity: By restructuring chromatin, SWI/SNF complexes facilitate the recruitment and activity of RNA polymerase and other transcriptional machinery at gene loci, promoting efficient gene. |
| Chromosomal Remodeling |
Nucleosome Repositioning: SWI/SNF complexes reposition nucleosomes along DNA, altering the packaging of DNA around histone proteins. This activity can lead to the displacement, sliding, or eviction of nucleosomes, regulating access to DNA sequences for transcriptional machinery. Histone Variant Exchange: SWI/SNF complexes can exchange histone variants within nucleosomes, influencing chromatin structure and gene expression patterns. This dynamic remodeling of chromatin plays a crucial role in regulating gene activity. |
| Epigenetics |
Interplay with Epigenetic Marks: SWI/SNF complexes interact with epigenetic modifications such as DNA methylation and histone acetylation or methylation. This crosstalk between chromatin remodeling and epigenetic marks influences gene expression by modulating chromatin accessibility and transcriptional activity. Regulation of DNA Accessibility: SWI/SNF complexes regulate the accessibility of DNA regions by modifying chromatin structure. This process affects the binding of transcription factors, RNA polymerase, and other regulatory proteins, ultimately controlling gene expression in response to cellular signals. |
| Cell Differentiation and Development |
Tissue-Specific Gene Expression: SWI/SNF complexes are crucial for regulating tissue-specific gene expression patterns during cell differentiation and development. By modulating chromatin structure, these complexes control the activation or repression of genes essential for cell fate determination and tissue morphogenesis. Maintenance of Stemness: SWI/SNF complexes play a role in maintaining stem cell identity by regulating the expression of genes associated with pluripotency and differentiation. Their activity is essential for the balance between self-renewal and differentiation in stem cell populations. Embryonic Development: SWI/SNF complexes are involved in embryonic development by regulating the expression of genes critical for patterning and organogenesis. Their activity is essential for proper development and differentiation of various cell lineages. |
| Cell Signaling and Response | Integration of Signals: SWI/SNF complexes integrate external signals and cellular cues to modulate gene expression in response to environmental changes. They play a role in transducing signals from the cell membrane to the nucleus, regulating gene transcription and cellular responses. |
In summary, the SWI/SNF family of chromatin remodeling complexes is integral to cell biology and biochemical processes by regulating transcriptional activity, remodeling chromatin structure, influencing epigenetic modifications, and orchestrating gene expression patterns critical for cell differentiation, development, and cellular homeostasis. Their multifaceted roles highlight the importance of SWI/SNF complexes in modulating gene expression and cellular function across various biological contexts.
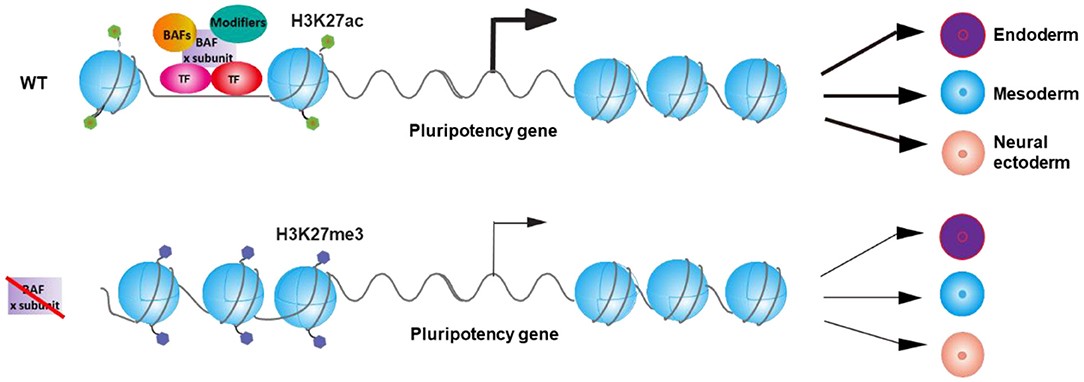 Fig.2 Model for the regulation of the balance between pluripotency and differentiation of embryonic stem cells (ESCs) by BAF complexes via the control of pluripotency gene expression. (Ye Y, et al., 2021)
Fig.2 Model for the regulation of the balance between pluripotency and differentiation of embryonic stem cells (ESCs) by BAF complexes via the control of pluripotency gene expression. (Ye Y, et al., 2021)
BAF complex, transcription factors, and other chromatin modifiers regulate the expression of specific pluripotency gene(s) and thereby control the balance between pluripotency and differentiation of ESCs. Inactivation of specific BAF subunits leads to the deregulation of the expression of specific pluripotency gene(s) and therefore results in the differential differentiation defects of ESC.
Members and Features of SWI/SNF Family
Here are some key members and features of the SWI/SNF family:
| Members | Features |
|---|---|
| SWI/SNF complex | The SWI/SNF complex is the founding member of this family. It is involved in altering nucleosome positioning and promoting access to DNA for various cellular processes, such as transcription, DNA repair, and recombination. |
| BRG1 (SMARCA4) | BRG1 is a catalytic subunit of the SWI/SNF complex. It possesses ATPase activity, which is essential for nucleosome remodeling and gene regulation. |
| BRM (SMARCA2) | BRM is another ATPase subunit of the SWI/SNF complex. It shares similarities with BRG1 and plays similar roles in chromatin remodeling and gene expression. |
| BAF complex | This is a mammalian SWI/SNF complex that consists of various subunits, including ARID1A, ARID1B, ARID2, BAF155, BAF170, and others. The BAF complex has distinct roles in different tissues and developmental stages. |
| PBAF complex | Another variant of the SWI/SNF complex, the PBAF (Polybromo-associated BAF) complex, contains additional subunits like BAF200, BAF180, and BRD7. It has specialized functions in gene regulation and genome stability. |
Overall, the SWI/SNF family of chromatin remodeling complexes is essential for the precise control of gene expression and plays diverse roles in development, differentiation, and disease.
The Association of SWI/SNF Family with Disease
Dysregulation of SWI/SNF complexes, often due to genetic mutations, is associated with various diseases, including cancer, neurodevelopmental disorders, and other conditions. Here are examples of diseases linked to SWI/SNF family dysfunction:
Cancer
- Loss of function mutations in SMARCB1, a core subunit of the SWI/SNF complex, are commonly found in malignant rhabdoid tumors. These aggressive pediatric malignancies are characterized by alterations in chromatin structure and uncontrolled cell growth.
- ARID1A, a subunit of the SWI/SNF complex, is frequently mutated in ovarian clear cell carcinoma. Dysregulation of ARID1A impacts chromatin remodeling and gene expression, contributing to tumorigenesis.
- Mutations in SMARCA4 have been implicated in non-small cell lung cancer (NSCLC). Alterations in BRG1 function can lead to changes in gene expression patterns and chromatin structure that promote tumor development.
Neurodevelopmental Disorders
- Mutations in ARID1B, a member of the SWI/SNF complex, have been associated with Coffin-Siris syndrome, a rare genetic disorder characterized by developmental delays and intellectual disability.
- Mutations in SMARCA2 have been linked to Nicolaides-Baraitser syndrome, a neurodevelopmental disorder featuring intellectual disability, seizures, and distinctive facial features. Disruption of SMARCA2 function affects chromatin remodeling critical for normal brain development.
Other Diseases
- Mutations in SMARCB1 have been associated with Schwannomatosis, a rare genetic condition characterized by the development of multiple schwannomas, and benign tumors affecting the peripheral nerves.
- Mutations in ARID1A have also been linked to Coffin-Siris-like syndrome, a disorder with features resembling Coffin-Siris syndrome but distinct clinical characteristics.
Dysfunction of the SWI/SNF family of chromatin remodeling complexes due to genetic mutations is associated with a range of diseases, including various cancers, neurodevelopmental disorders, and other conditions. Understanding how alterations in SWI/SNF complex components impact gene regulation, chromatin structure, and cellular processes is crucial for elucidating disease mechanisms and developing targeted therapeutic strategies for these disorders.
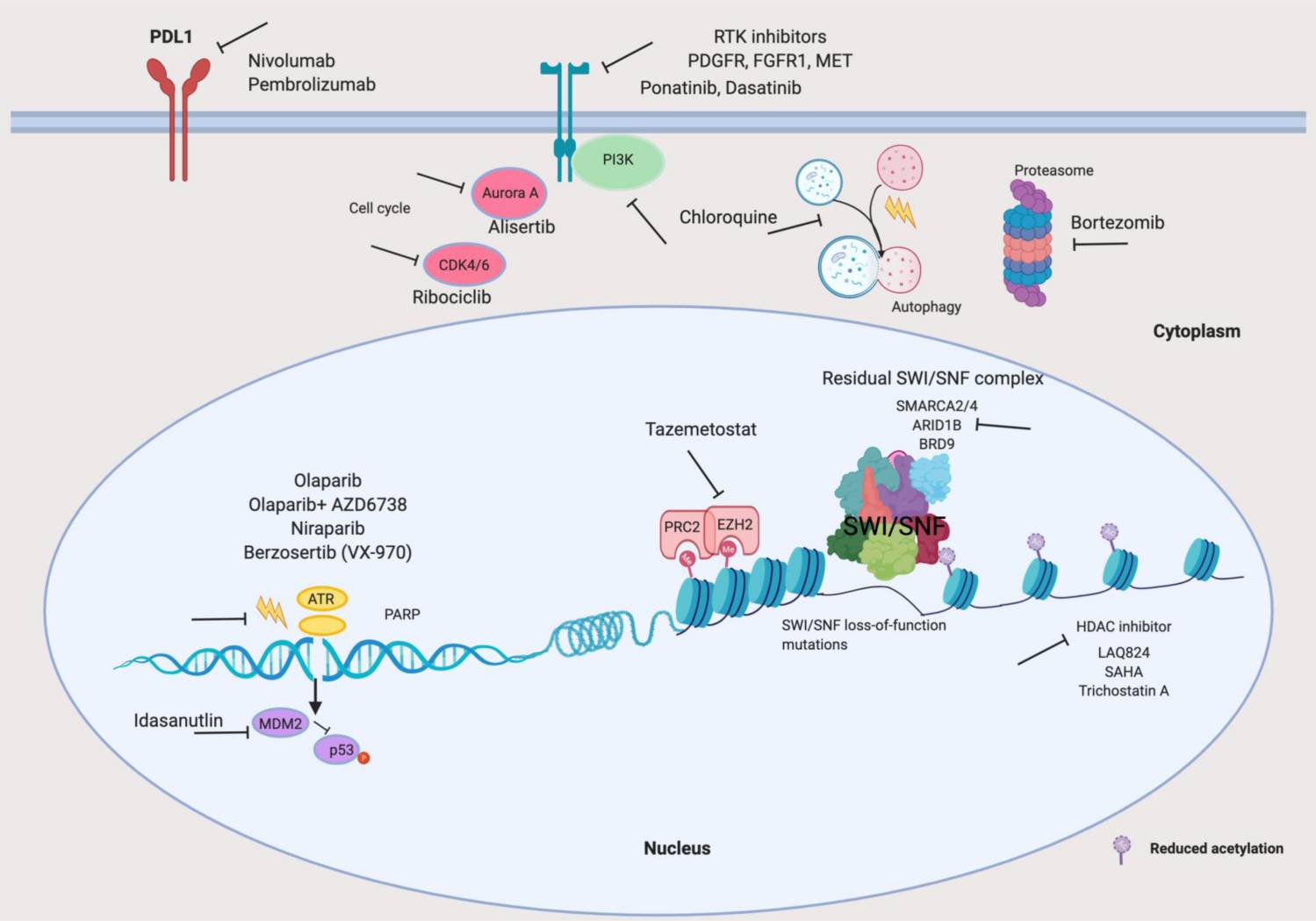 Fig.3 Translational science of cancers with SWI/SNF complex aberrations. (Mittal P, et al., 2020)
Fig.3 Translational science of cancers with SWI/SNF complex aberrations. (Mittal P, et al., 2020)Case Study
Case 1: Wang Y, Hoang L, Ji JX, Huntsman DG. SWI/SNF complex mutations in gynecologic cancers: molecular mechanisms and models. Annu Rev Pathol. 2020;15:467-492.
The SWI/SNF chromatin remodeling complexes play a crucial role in regulating gene expression by interacting with histones and transcription factors to modulate chromatin structure. These evolutionary conserved multi-subunit protein complexes are essential for various biological functions, including differentiation and cell proliferation. Recent genomic studies have uncovered frequent mutations in genes encoding SWI/SNF complex subunits across a broad range of cancer types, particularly in gynecologic cancers. These mutations manifest at different tumor development stages and are specific to distinct histologic subtypes of gynecologic cancer. Therefore, the impact of SWI/SNF mutations on gynecologic cancer initiation and progression is contingent upon the specific tissue and cellular context. This review aims to consolidate current knowledge on SWI/SNF mutations in the context of gynecologic cancer development, shedding light on the molecular mechanisms and potential therapeutic implications for these diseases.
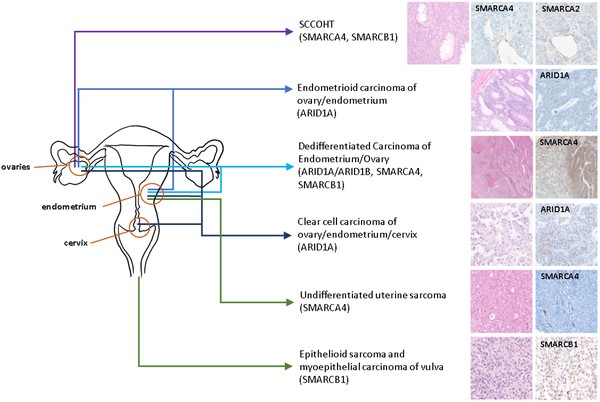 Fig.1 SWI/SNF mutations and protein loss in gynecologic cancers.
Fig.1 SWI/SNF mutations and protein loss in gynecologic cancers.Case 2: Smith JJ, Xiao Y, Parsan N, et al. The SWI/SNF chromatin remodeling assemblies BAF and PBAF differentially regulate cell cycle exit and cellular invasion in vivo. PLoS Genet. 2022;18(1):e1009981.
In their study, researchers explored how chromatin remodelers, notably the SWI/SNF complex, orchestrate metazoan development by finely regulating chromatin accessibility and transcription. This meticulous control ensures proper cell cycle management and differentiation in a specific lineage and time-dependent manner. Mutations in genes responsible for chromatin's structural subunits, like histones, and factors regulating chromatin function are implicated in various disease processes, including cancer metastasis.
Using Caenorhabditis elegans anchor cell (AC) invasion as a model system, the researchers identified a range of chromatin agents and regulators that drive cellular invasiveness. They found that the SWI/SNF ATP-dependent chromatin remodeling complex plays a crucial role in regulating AC invasion, influencing both G0 cell cycle arrest and the activation of invasive mechanisms. Through targeted protein degradation and enhanced RNA interference techniques, the study demonstrated that SWI/SNF modulates AC invasion in a dosage-dependent manner. Lower SWI/SNF activity in the AC resulted in irregular cell cycle progression and increased invasion failure.
Their data specifically linked the SWI/SNF BAF assembly to G0 cell cycle arrest regulation in the AC, while the SWI/SNF PBAF assembly promoted AC invasion through cell cycle-independent processes, such as interactions with the basement membrane and activation of pro-invasive genes. These findings underscore the essential role of the SWI/SNF complex in two critical aspects of AC invasion: halting cell cycle progression and restructuring the basement membrane. This research offers valuable insights into how distinct SWI/SNF assemblies contribute differentially to cellular invasion and how disruptions in specific SWI/SNF subunits may contribute to tumor development and cancer spread.
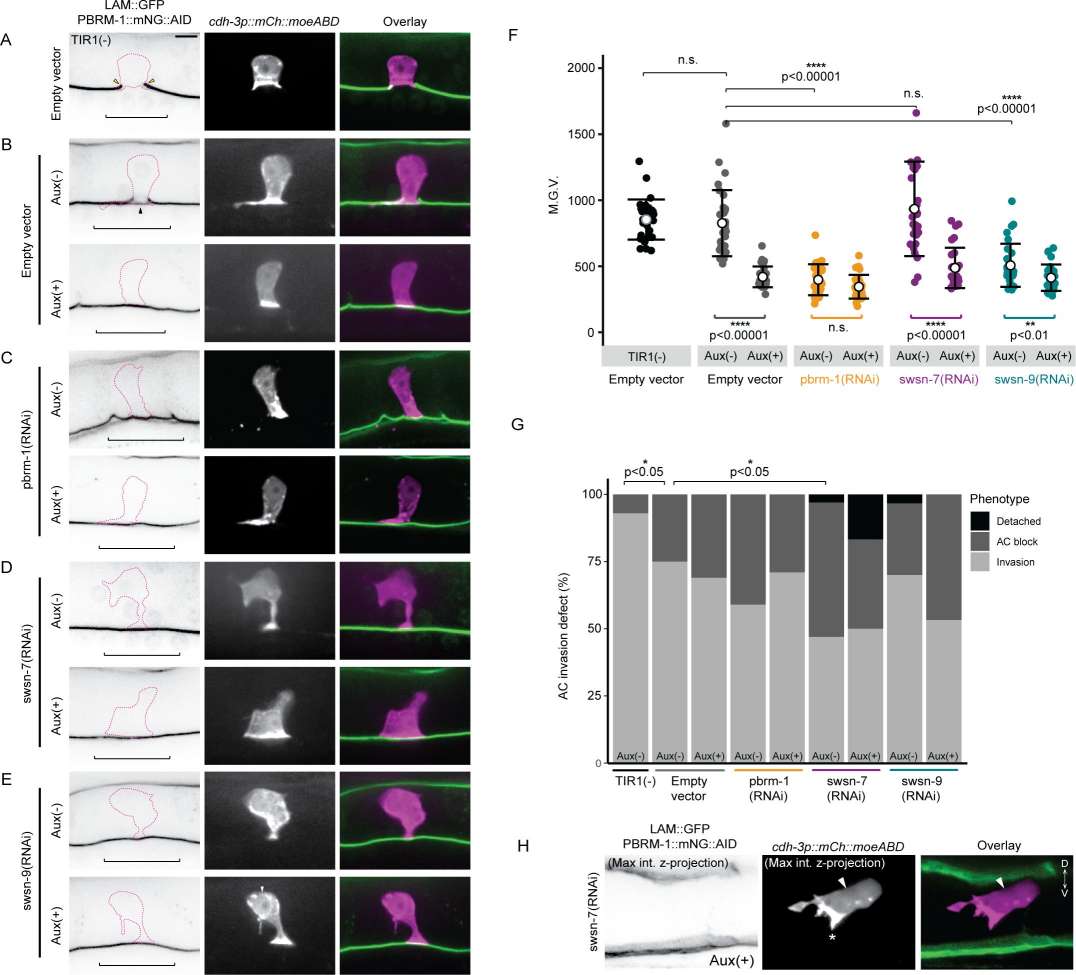 Fig.2 PBAF promotes anchor cell (AC) contact to the basement membrane (BM).
Fig.2 PBAF promotes anchor cell (AC) contact to the basement membrane (BM).Related References
- Chen K, Yuan J, Sia Y, Chen Z. Mechanism of action of the SWI/SNF family complexes. Nucleus. 2023;14(1):2165604.
- Wang Y, Hoang L, Ji JX, Huntsman DG. SWI/SNF complex mutations in gynecologic cancers: molecular mechanisms and models. Annu Rev Pathol. 2020;15:467-492.
- Ye Y, Chen X, Zhang W. Mammalian SWI/SNF chromatin remodeling complexes in embryonic stem cells: regulating the balance between pluripotency and differentiation. Front Cell Dev Biol. 2021;8:626383.
- Mittal P, Roberts CWM. The SWI/SNF complex in cancer - biology, biomarkers and therapy. Nat Rev Clin Oncol. 2020;17(7):435-448.
