Histone Demethylases
Related Symbol Search List
Immunology Background
Background
Histone demethylases are a group of enzymes that play a crucial role in the dynamic regulation of gene expression by removing methyl groups from histone proteins. Histone demethylation is an essential process in epigenetic control, as it modulates the structure of chromatin and influences gene activity. There are two major families of histone demethylases: the amine oxidase-like proteins (such as KDM1/LSD1) and the Jumonji C (JmjC) domain-containing proteins.
Different Types of Histone Demethylases and Their Characteristics and Functions
| Types | Details |
|---|---|
| Amine oxidase-like proteins (KDM1/LSD1) |
Structure: Amine oxidase-like proteins, such as lysine-specific demethylase 1 (LSD1) or KDM1A, contain a unique amine oxidase-like domain. Substrate specificity: KDM1/LSD1 primarily acts on mono- and dimethylated lysine residues, particularly lysine 4 (K4) and lysine 9 (K9) on histone H3. Co-factors: KDM1/LSD1 requires flavin adenine dinucleotide (FAD) as a cofactor for its demethylase activity. Function: KDM1/LSD1 plays a crucial role in transcriptional regulation and development. It functions as a component of large protein complexes involved in gene activation or repression. KDM1/LSD1 is involved in the regulation of various biological processes, including cell differentiation, embryonic development, and cancer progression. |
| Jumonji C (JmjC) domain-containing proteins |
Structure: JmjC domain-containing proteins make up the largest family of histone demethylases. They possess a conserved JmjC domain involved in catalytic activity. Substrate specificity: JmjC domain-containing proteins exhibit diverse substrate specificities, targeting various lysine and arginine residues on histones. Different members of this family can demethylate mono-, di-, or trimethylated lysine or arginine residues. Co-factors: JmjC domain-containing demethylases require alpha-ketoglutarate (α-KG) and molecular oxygen as cofactors for their demethylase activity. Function: JmjC domain-containing proteins are involved in a wide range of biological processes, including development, differentiation, and disease. They play roles in gene regulation, DNA repair, and cellular signaling pathways. Examples of JmjC domain-containing demethylases include KDM2-7, KDM4, KDM5, and KDM6 subfamilies, each with their specific functions and target sites. |
Mechanism of Action of Histone Demethylases
Histone demethylases are enzymes that catalyze the removal of methyl groups from histone proteins, thereby reversing the methylation marks on specific lysine or arginine residues. This process is essential for the dynamic regulation of gene expression and the maintenance of proper chromatin structure. The mechanism of action of histone demethylases involves several steps:
- Recognition of methylated histones: Histone demethylases possess specific domains or motifs that can recognize and bind to methylated histones. These domains allow demethylases to target specific methylated sites on histones.
- Demethylation reaction: Once bound to the methylated histones, histone demethylases catalyze the removal of methyl groups from the targeted lysine or arginine residues. There are two main mechanisms involved in demethylation:
a. Oxidative demethylation: Some histone demethylases, such as the amine oxidase-like proteins (e.g., KDM1/LSD1), utilize an oxidative mechanism. They use flavin adenine dinucleotide (FAD) as a cofactor and generate reactive oxygen species (ROS) to oxidize the methyl group on histones. This process results in the release of formaldehyde as a byproduct.
b. Hydroxylation demethylation: Another group of histone demethylases, known as Jumonji C (JmjC) domain-containing demethylases, employ a hydroxylation-based mechanism. These enzymes use alpha-ketoglutarate (α-KG) and molecular oxygen as cofactors. The demethylation reaction involves hydroxylation of the methylated lysine or arginine residue, followed by the removal of the hydroxylated group. In this process, succinate is produced as a byproduct. - Co-factors and regulation: Histone demethylases require specific co-factors for their activity. These co-factors include FAD, α-KG, and molecular oxygen. The availability and interaction of these co-factors can influence the activity and specificity of histone demethylases. Additionally, histone demethylases can be regulated by various mechanisms, including protein-protein interactions, post-translational modifications, and subcellular localization.
By removing methyl groups from histones, histone demethylases contribute to the dynamic regulation of gene expression. The removal of methyl marks can lead to changes in chromatin structure, allowing for the activation or repression of specific genes. The precise targeting and activity of histone demethylases are critical for maintaining proper gene expression patterns and cellular functions.
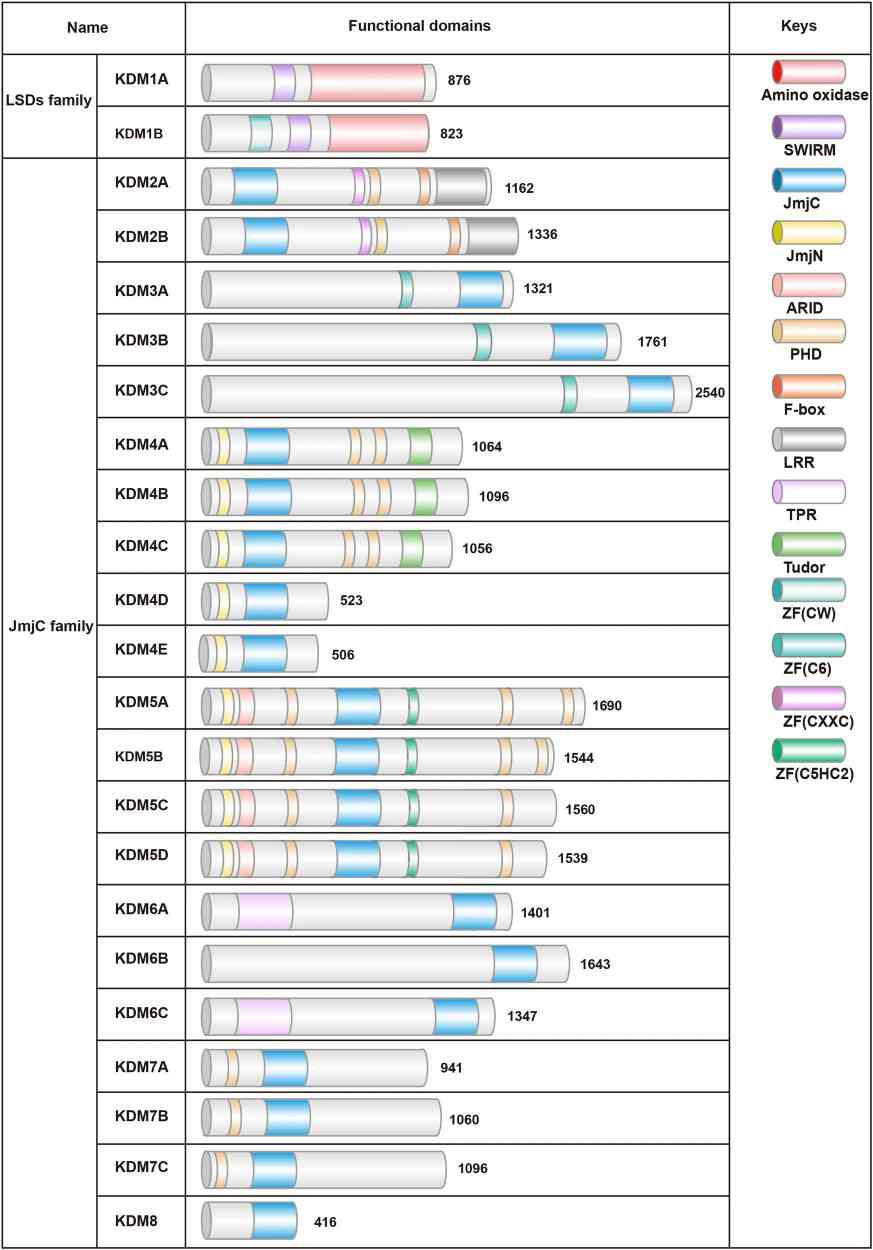 Fig.1 Schematic diagram of two families of human histone demethylases (KDMs). (Qu L, et al., 2023)
Fig.1 Schematic diagram of two families of human histone demethylases (KDMs). (Qu L, et al., 2023)Representative Members of Histone Demethylases
| Members | Function |
|---|---|
| LSD1/KDM1A | It is the founding member of the amine oxidase-like proteins. LSD1 primarily targets mono- and dimethylated lysine residues, particularly lysine 4 (K4) and lysine 9 (K9) on histone H3. It plays a critical role in various biological processes, including development, differentiation, and cancer progression. |
| KDM2A/FBXL11 | KDM2A is a JmjC domain-containing demethylase that primarily targets dimethylated lysine 36 (K36) on histone H3 (H3K36me2). It is involved in transcriptional regulation, X-chromosome inactivation, and DNA damage response pathways. |
| KDM4A/JMJD2A | KDM4A is a member of the KDM4 subfamily of JmjC domain-containing demethylases. It demethylates trimethylated lysine 9 and lysine 36 on histone H3 (H3K9me3 and H3K36me3). KDM4A is implicated in gene regulation, DNA repair, and cellular senescence. |
| KDM6A/UTX | KDM6A is a well-known JmjC domain-containing demethylase that targets trimethylated lysine 27 on histone H3 (H3K27me3). It is involved in gene activation, development, X-chromosome inactivation, and plays a tumor suppressor role in various cancers. |
| KDM5C/JARID1C | KDM5C belongs to the KDM5 subfamily of JmjC domain-containing demethylases. It specifically targets di- and trimethylated lysine 4 on histone H3 (H3K4me2/3). Mutations in KDM5C are associated with intellectual disability and X-linked syndromic mental retardation. |
These representative members exemplify the diversity and functional significance of histone demethylases. Each member of the family possesses unique characteristics and targets specific methyl marks on histones, contributing to the dynamic regulation of gene expression and chromatin structure.
The Key Role of Histone Demethylases
Histone demethylases play critical roles in cell cycle regulation, gene expression, and epigenetics. Here's an overview of their involvement in these processes:
| The key roles | Details |
|---|---|
| Cell Cycle Regulation |
|
| Gene Expression |
|
| Epigenetics |
|
Histone demethylases exhibit substrate specificity and target specific methyl marks on histones to modulate chromatin structure, gene accessibility, and transcriptional outcomes. Dysregulation of histone demethylases can disrupt these processes and contribute to various diseases, underscoring their importance in maintaining proper cellular function and homeostasis.
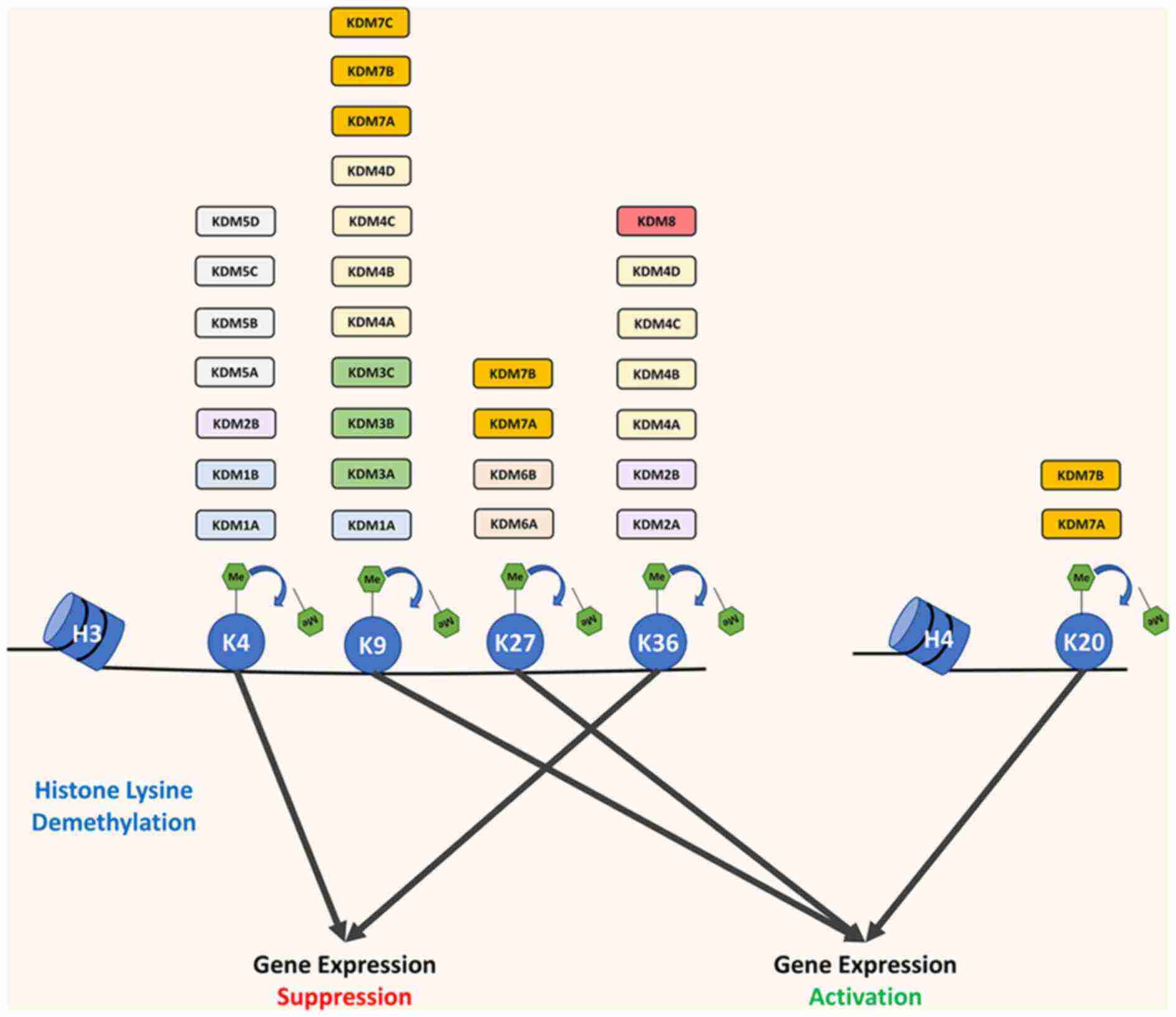 Fig.2 Overview of histone demethylases targeting the site of histone lysine and their effect on gene expression. (Diao W, et al., 2022)
Fig.2 Overview of histone demethylases targeting the site of histone lysine and their effect on gene expression. (Diao W, et al., 2022)Role of Histone Demethylases in Diseases
Histone demethylases play critical roles in the occurrence and development of various diseases. Dysregulation of these enzymes can lead to aberrant gene expression patterns and contribute to disease pathogenesis. Here are examples of the involvement of histone demethylases in specific diseases:
Cancer
Histone demethylases have been extensively implicated in different types of cancers. Dysregulation of these enzymes can lead to the activation or repression of specific genes involved in cancer development, progression, and metastasis. Histone demethylases can either act as oncogenes or tumor suppressors depending on the specific context and target genes involved.
- Overexpression of LSD1 has been observed in several cancers, including leukemia, prostate cancer, and breast cancer. LSD1 promotes tumorigenesis by repressing tumor suppressor genes and activating oncogenes. Its dysregulation is associated with cancer progression, metastasis, and drug resistance.
- KDM4C/JMJD2C is frequently upregulated in various cancers, including breast, lung, and colorectal cancers. It promotes cancer cell proliferation, invasion, and metastasis by regulating the expression of genes involved in cell cycle control and epithelial-mesenchymal transition (EMT).
Neurological Disorders
Histone demethylases are involved in neuronal development, synaptic plasticity, and cognitive processes. Dysregulation of these enzymes has been associated with several neurological disorders, including neurodevelopmental disorders, neurodegenerative diseases, and psychiatric conditions.
- KDM5A/JARID1A has been implicated in neurodevelopmental disorders, such as autism spectrum disorders (ASDs). Dysregulation of KDM5A affects the expression of genes involved in neuronal development and synaptic function, contributing to the pathogenesis of ASDs.
- LSD1/KDM1A is involved in neurodegenerative diseases, including Alzheimer's disease. Its dysregulation affects the expression of genes associated with neuroinflammation, beta-amyloid accumulation, and synaptic dysfunction, contributing to disease progression.
Cardiovascular Diseases
Histone demethylases play a role in cardiovascular development and disease progression. The dysregulated activity of these enzymes can contribute to cardiac hypertrophy, fibrosis, and other cardiovascular disorders by affecting gene expression patterns involved in cardiac remodeling and function.
- KDM4B/JMJD2B has been implicated in cardiac hypertrophy and heart failure. Its overexpression promotes pathological cardiac remodeling by regulating gene expression involved in cardiac hypertrophy and fibrosis.
- KDM6A/UTX mutations are associated with Kabuki syndrome, a congenital disorder characterized by multiple congenital anomalies, including cardiovascular defects.
Developmental Disorders
Proper regulation of histone methylation is essential for normal development. Dysregulation of histone demethylases during embryonic development or early stages of life can disrupt gene expression patterns critical for proper organogenesis and tissue differentiation, leading to developmental disorders.
- Dysregulation of (LSD1)/KDM1A has been linked to developmental disorders, such as Rett syndrome. LSD1 plays a role in neuronal development and maturation, and its abnormal activity can affect gene expression patterns critical for normal brain development.
Immune Disorders
- Histone demethylases participate in the regulation of immune responses and inflammatory processes. Dysregulation of these enzymes can impact immune cell differentiation, activation, and cytokine production, leading to immune disorders or chronic inflammation.
These examples highlight the involvement of histone demethylases in various diseases by influencing gene expression patterns and chromatin states. Dysregulation of these enzymes can disrupt normal cellular processes, contribute to disease pathogenesis, and serve as potential therapeutic targets for intervention in the future. It is important to note that the specific roles of histone demethylases in diseases are still an active area of research, and further studies are needed to fully elucidate their mechanisms and therapeutic implications.
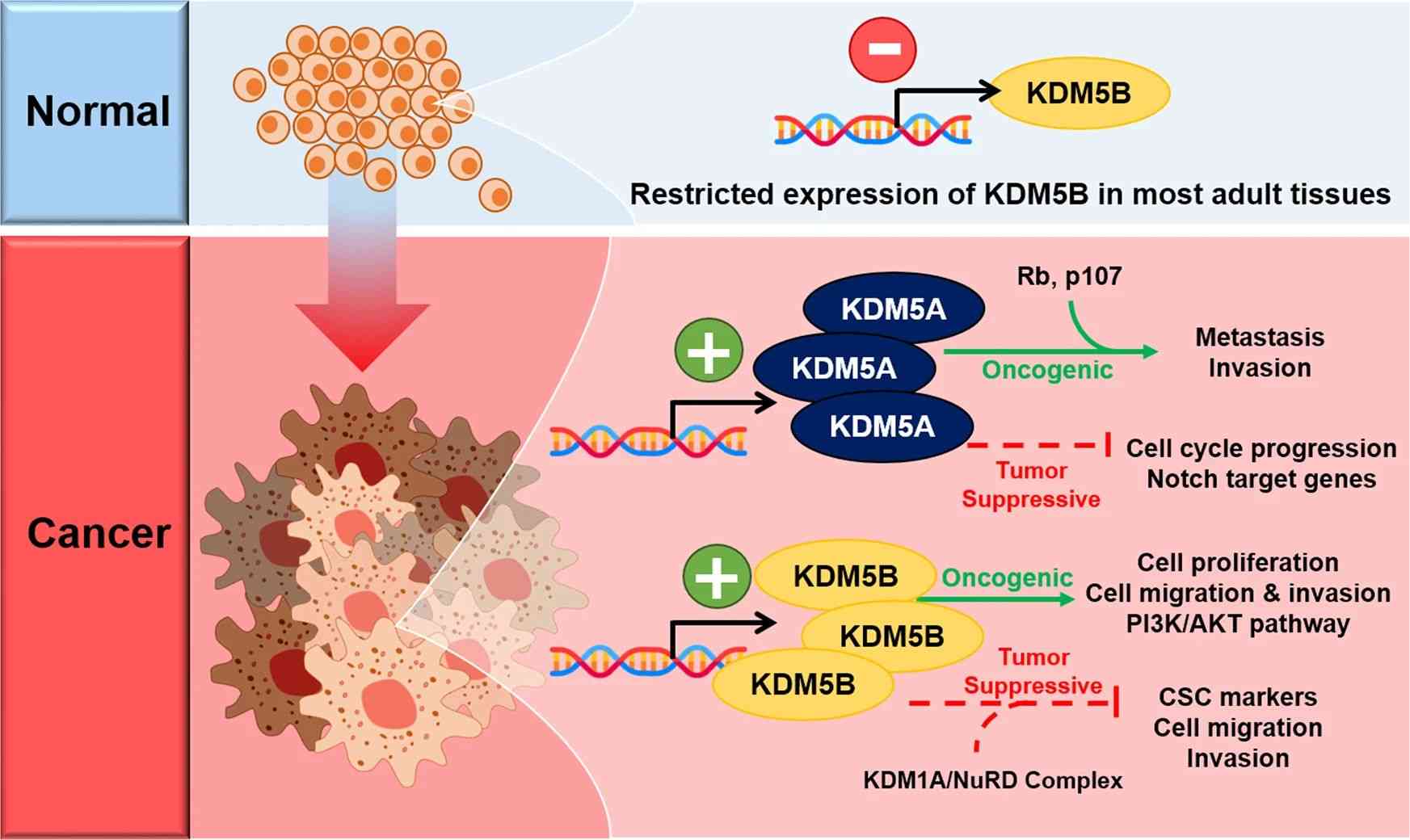 Fig.3 Context-dependent oncogenic and tumor-suppressive roles of KDM5A and KDM5B in breast and prostate tumorigenesis. (Yoo J, et al., 2022)
Fig.3 Context-dependent oncogenic and tumor-suppressive roles of KDM5A and KDM5B in breast and prostate tumorigenesis. (Yoo J, et al., 2022)Case Study
Case 1: Lin C-Y, Wang B-J, Chen B-C, Tseng J-C, Jiang SS, Tsai KK, Shen Y-Y, Yuh CH, Sie Z-L, Wang W-C, et al. Histone demethylase KDM4C stimulates the proliferation of prostate cancer cells via activation of AKT and c-Myc. Cancers. 2019; 11(11):1785.
The study demonstrated that the histone demethylase KDM4C promotes the proliferation of prostate cancer (PCa) cells. The findings showed that KDM4C hindered the acini morphogenesis of prostate cells and its knockdown suppressed cell proliferation and tumor growth. Micro-Western array analysis revealed that KDM4C knockdown decreased the phosphorylation of AKT, c-Myc, and other proteins involved in cell cycle regulation, while increasing the expression of PTEN. KDM4C was found to co-localize with AR and c-Myc in the nuclei of PCa cells. Overexpression of AKT or c-Myc rescued the suppressive effect of KDM4C knockdown on PCa cell proliferation. Moreover, the expression of KDM4C mRNA and protein was higher in prostate tumor tissues compared to adjacent normal prostate tissues, and higher KDM4C protein expression correlated with higher levels of AKT and c-Myc proteins. These findings suggest that KDM4C promotes PCa cell proliferation through the activation of c-Myc and AKT.
The study also examined clinical samples and found that KDM4C mRNA expression was higher in prostate tumors compared to normal prostate tissues in multiple datasets. Immunohistochemical staining further confirmed higher protein expression of KDM4C, AKT, and c-Myc in prostate carcinoma tissues compared to adjacent normal prostate tissues, indicating a positive correlation between KDM4C expression and the expression of AKT and c-Myc in prostate tumors.
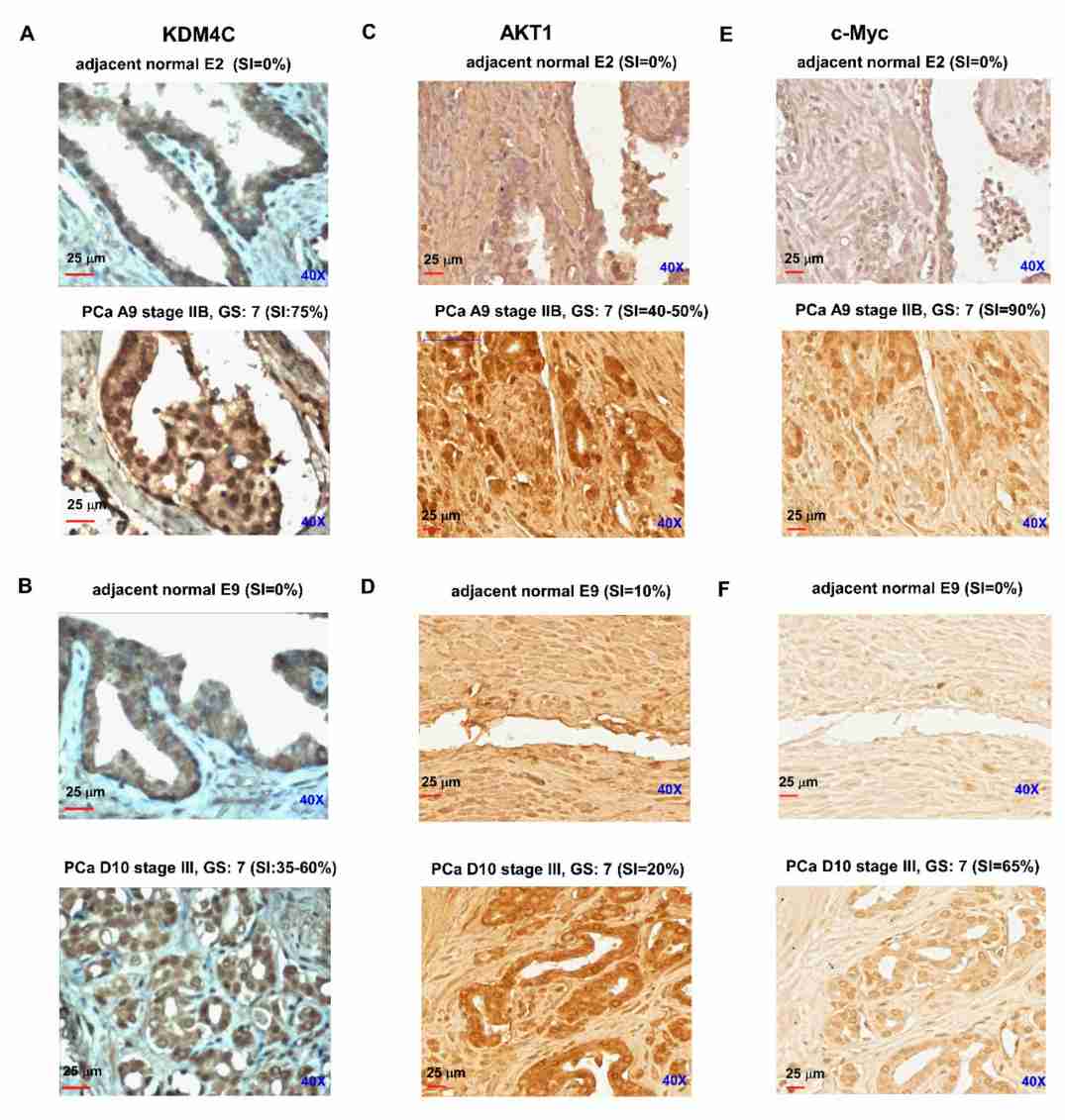 Fig.1 Protein expression level of KDM4C in prostate carcinoma versus adjacent normal prostate tissues.
Fig.1 Protein expression level of KDM4C in prostate carcinoma versus adjacent normal prostate tissues.Case 2: Chen D, Cai B, Zhu Y, et al. Targeting histone demethylases JMJD3 and UTX: selenium as a potential therapeutic agent for cervical cancer. Clin Epigenetics. 2024;16(1):51.
Selenium has shown remarkable inhibitory effects on cervical cancer cells in various ways. One of the mechanisms behind selenium's inhibitory effects is its ability to inhibit histone demethylases, specifically JMJD3 and UTX. This inhibition is observed both in vitro and in vivo. Notably, when JMJD3 and UTX are inhibited with GSK-J4, similar biological effects are observed in both in vitro and in vivo models, effectively inhibiting organoid models derived from cervical cancer patients. Inhibiting JMJD3 and UTX also induces G2/M phase arrest, promotes cellular apoptosis, and reverses epithelial-mesenchymal transition (EMT). ChIP-qPCR analysis confirms that JMJD3 and UTX inhibition increases the recruitment of a specific histone modification, H3K27me3, to the transcription start sites (TSS) of target genes in cervical cancer cells (HeLa and SiHa cells). Furthermore, the expressions of JMJD3 and UTX are found to be significantly higher in cervical cancer tissues compared to adjacent normal cervical tissues, suggesting their potential as therapeutic targets.
The study investigated the role of histone H3K27 demethylases JMJD3 and UTX in cervical cancer using the specific inhibitor GSK-J4. The results showed that GSK-J4 treatment significantly inhibited cell viability and impaired the colony formation ability of HeLa and SiHa cervical cancer cells. Flow cytometry analysis revealed that GSK-J4 treatment led to an increase in the percentage of cells in the G2/M phase and induced apoptosis in both cell lines. qRT-PCR and Western blot analysis demonstrated that histone H3K27 demethylase inhibition downregulated the expression of cell cycle-related genes (Cyclin B1 and CDK1) and apoptosis-related genes (Bcl-2 and Ki67), while upregulating the expression of pro-apoptotic genes (Bax, caspase-7, and cleaved caspase-3). The study provides evidence for the involvement of JMJD3 and UTX in cervical cancer cell proliferation, cell cycle progression, and apoptosis, suggesting their potential as therapeutic targets in cervical cancer treatment.
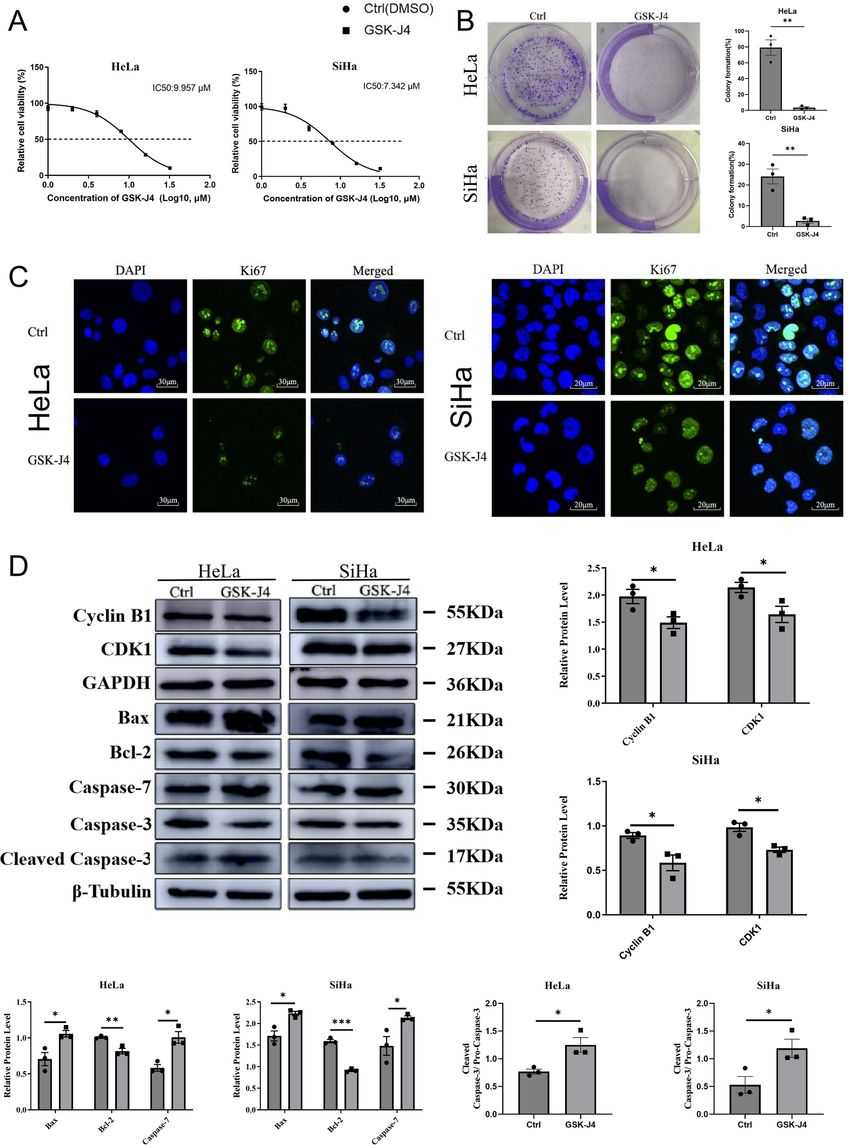 Fig.2 The effects of inhibiting histone H3K27 demethylases on the proliferation of cervical cancer cells.
Fig.2 The effects of inhibiting histone H3K27 demethylases on the proliferation of cervical cancer cells.Related References
- Dimitrova E, Turberfield AH, Klose RJ. Histone demethylases in chromatin biology and beyond. EMBO Rep. 2015;16(12):1620-1639.
- Zhang Y, Chen J, Liu H, et al. The role of histone methylase and demethylase in antitumor immunity: A new direction for immunotherapy. Front Immunol. 2023;13:1099892. Published 2023 Jan 11.
- Sterling J, Menezes SV, Abbassi RH, Munoz L. Histone lysine demethylases and their functions in cancer. Int J Cancer. 2021;148(10):2375-2388.
- Qu, L., Yin, T., Zhao, Y. et al. Histone demethylases in the regulation of immunity and inflammation. Cell Death Discov. 9, 188 (2023).
- Diao W, Zheng J, Li Y, Wang J, Xu S. Targeting histone demethylases as a potential cancer therapy (Review). Int J Oncol. 2022;61(3):103.
- Yoo J, Kim GW, Jeon YH, Kim JY, Lee SW, Kwon SH. Drawing a line between histone demethylase KDM5A and KDM5B: their roles in development and tumorigenesis. Exp Mol Med. 2022;54(12):2107-2117.
