Histone Methyltransferase
Related Symbol Search List
Immunology Background
Background
Histone methyltransferases (HMTs) are enzymes that catalyze the addition of methyl groups to histone proteins. These enzymes play a crucial role in the epigenetic regulation of gene expression by modifying the structure of chromatin.
Structural Characteristics and Mechanism of Catalyzing Histone Methylation Modification
HMTs exhibit specific structural characteristics that enable them to catalyze methylation modifications on histones. Here are the key structural features and the catalytic mechanism of HMTs:
- SET Domain: The SET domain is a conserved structural motif found in most HMTs. It is responsible for the catalytic activity of transferring methyl groups to histone substrates. The SET domain consists of around 130 amino acids and contains the catalytic site where the methylation reaction takes place.
- Structural Variations: HMTs can display structural variations based on their specific functions and substrate specificities. Some HMTs contain additional domains or regions that contribute to substrate recognition, protein-protein interactions, or regulation of their catalytic activity. Examples include the post-SET domain, pre-SET domain, and methyltransferase-associated domains (SRA or Tudor domains).
- Substrate Recognition: HMTs have specific substrate preferences and recognize particular histone residues for methylation. The substrate specificity is determined by the amino acid sequence and structural features of the SET domain and its flanking regions. Different HMTs can target specific lysine or arginine residues on histones for methylation.
- Co-factor Binding: HMTs require a co-factor, typically S-adenosylmethionine (SAM), as the methyl group donor. SAM binds to the active site of the SET domain, providing the methyl group for transfer during the methylation reaction.
Methyl Transfer Mechanism: The catalytic mechanism of HMTs involves several steps:
- Substrate Binding: The histone substrate binds to the active site of the SET domain, positioning the target residue for methylation.
- Methyl Group Transfer: SAM, the co-factor, donates a methyl group to the target histone residue. This methyl transfer reaction is facilitated by the catalytic residues within the SET domain.
- Product Release: After the methyl group is transferred, the modified histone and the byproduct S-adenosylhomocysteine (SAH) are released from the active site.
Overall, the structural characteristics of HMTs, particularly the presence of the SET domain, enable their catalytic activity in transferring methyl groups to specific histone residues. The substrate recognition, co-factor binding, and methyl transfer mechanism are key aspects of histone methylation catalyzed by HMTs. Understanding the structural features and catalytic mechanisms of HMTs is crucial for deciphering the complex epigenetic regulation of gene expression and its implications in various biological processes and diseases.
Function
HMTs play a critical role in various cellular processes, including gene expression, chromatin remodeling, development, and disease. The addition of methyl groups to specific lysine or arginine residues on histones can lead to either gene activation or repression, depending on the context and the specific site of methylation. This histone methylation pattern serves as a binding platform for various effector proteins, including chromatin remodeling complexes and transcription factors, which modulate gene expression and chromatin structure.
The Different Subtypes and Members of Histone Methyltransferase
HMTs constitute a diverse group of enzymes, and they can be classified into several subtypes based on their structural features, catalytic activities, and functional roles. Here are some prominent subtypes of HMTs and their roles in different biological processes:
| Subtypes | Details |
|---|---|
| SET Domain-Containing HMTs |
EZH2 (Enhancer of Zeste Homolog 2) is a key component of the Polycomb Repressive Complex 2 (PRC2) and catalyzes the trimethylation of histone H3 at lysine 27 (H3K27me3). This modification is associated with gene repression and plays a critical role in embryonic development, stem cell maintenance, and cancer progression. SETD2 (SET Domain-Containing 2) is responsible for methylating histone H3 at lysine 36 (H3K36me3). This modification is associated with transcriptional elongation and DNA repair processes, ensuring genome stability. SETD8 catalyzes the monomethylation of histone H4 at lysine 20 (H4K20me1). It is involved in the regulation of DNA damage repair, genomic stability, and cell cycle progression. SETD7 catalyzes the monomethylation of histone H3 at lysine 4 (H3K4me1). It is associated with transcriptional regulation, enhancer activity, and chromatin organization. SUV39H1 and SUV39H2 are both SET domain-containing methyltransferases that catalyze the trimethylation of histone H3 at lysine 9 (H3K9me3). They are involved in heterochromatin formation, gene silencing, and maintenance of genomic stability. |
| Dot1-Like HMTs | DOT1L (Disruptor Of Telomeric Silencing 1-Like): DOT1L catalyzes the methylation of histone H3 at lysine 79 (H3K79me2/me3). H3K79 methylation is implicated in gene activation, DNA damage response, and leukemogenesis. |
| PRMTs (Protein Arginine Methyltransferases) | PRMT5 methylates histone H4 at arginine 3 (H4R3me2) and histone H3 at arginine 2 (H3R2me2). These modifications are involved in gene repression and are associated with diverse cellular processes such as transcriptional regulation, RNA processing, and stem cell maintenance. |
| PRDM Proteins (PRDI-BF1 and RIZ Domain-Containing Proteins) | PRDM2 is involved in the methylation of histone H3 at lysine 9 (H3K9me1/me2) and contributes to transcriptional repression and the regulation of cell cycle progression. |
| G9a/GLP (EHMT2 and EHMT1) | G9a (EHMT2) and GLP (EHMT1) are responsible for the di- and trimethylation of histone H3 at lysine 9 (H3K9me2/me3). H3K9 methylation is associated with gene silencing, heterochromatin formation, and genomic imprinting. |
| SET and MYND Domain-Containing Protein | SMYD2 methylates histone H3 at lysine 36 (H3K36me1/me2). It is involved in gene transcription, DNA repair, and cell cycle regulation. |
These are just a few examples of histone methyltransferase subtypes and their roles in biological processes. However, it is important to note that many HMTs exhibit overlapping functions, and their specific roles can vary depending on the cellular context and the target genes they regulate. The complex interplay between different HMTs and their modifications contributes to the dynamic regulation of gene expression, chromatin structure, and various biological processes.
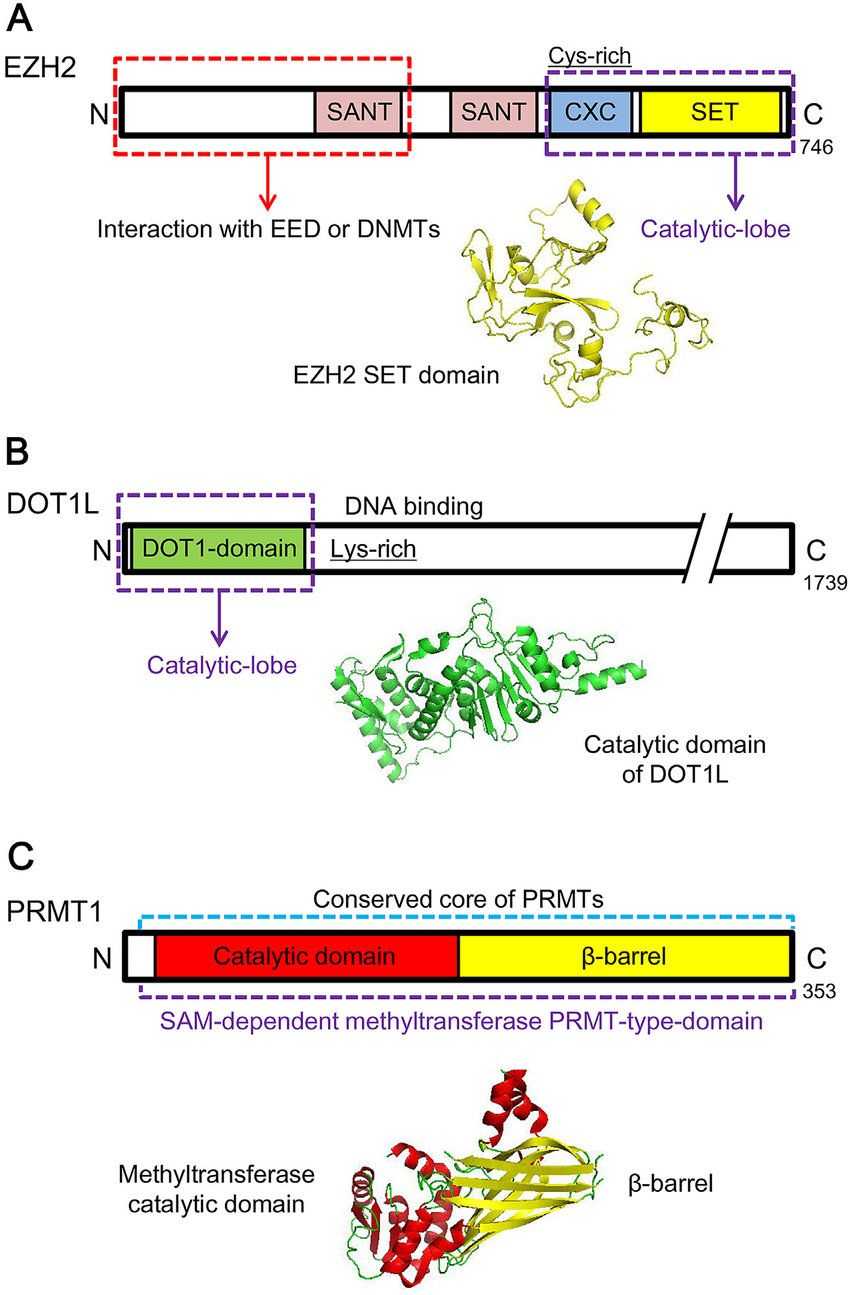 Fig.1 Schematic representations and structures of representative histone methylation 'writers'. (Chen Y, et al., 2018)
Fig.1 Schematic representations and structures of representative histone methylation 'writers'. (Chen Y, et al., 2018)The Abnormal Expression and Functional Changes of HMTs in Diseases
Abnormal expression and functional changes of HMTs have been observed in various diseases. Dysregulation of HMTs can lead to altered gene expression patterns, disrupted chromatin structure, and contribute to disease pathogenesis. Here are some examples of diseases where aberrant HMT expression or activity has been implicated:
Cancer
- Increased expression or gain-of-function mutations in EZH2 have been observed in several cancer types, including lymphomas, breast cancer, and prostate cancer. Elevated EZH2 activity leads to aberrant trimethylation of H3K27 (H3K27me3), resulting in gene silencing and promoting tumor growth and metastasis.
- Reduced expression of SUV39H1 and SUV39H2 has been associated with various cancers, including colorectal, lung, and breast cancer. Decreased H3K9 trimethylation (H3K9me3) mediated by SUV39H1/2 is linked to loss of heterochromatin and genomic instability.
- Loss-of-function mutations in SETD2 have been identified in renal cell carcinoma and other cancers. SETD2 is involved in H3K36 methylation (H3K36me3), and its disruption results in altered gene expression patterns and defective DNA repair.
Neurological Disorders
- Aberrant DOT1L activity has been implicated in neurodevelopmental disorders and acute leukemias. Inhibition of DOT1L has shown potential therapeutic effects in treating mixed lineage leukemia (MLL)-rearranged leukemia.
- Reduced expression of PRDM2 and subsequent loss of H3K9 methylation (H3K9me2) have been observed in neuroblastoma and glioblastoma. PRDM2 functions as a tumor suppressor by regulating cell cycle progression and apoptosis.
Cardiovascular Diseases
- Altered expression of SETD7 has been associated with cardiovascular diseases, including atherosclerosis and cardiac hypertrophy. SETD7-mediated H3K4 monomethylation (H3K4me1) plays a role in regulating gene expression during cardiac development and disease progression.
Immunological Disorders
- Aberrant EZH2 expression has been implicated in autoimmune diseases, such as systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA). Overexpression of EZH2 contributes to abnormal gene silencing and dysregulated immune responses.
These examples highlight the involvement of histone methyltransferases in various diseases. However, it's important to note that the roles of HMTs in disease are complex and context-dependent, and further research is needed to fully understand their contributions to specific pathological conditions. Targeting HMTs and their associated modifications is an active area of investigation for the development of potential therapeutic strategies.
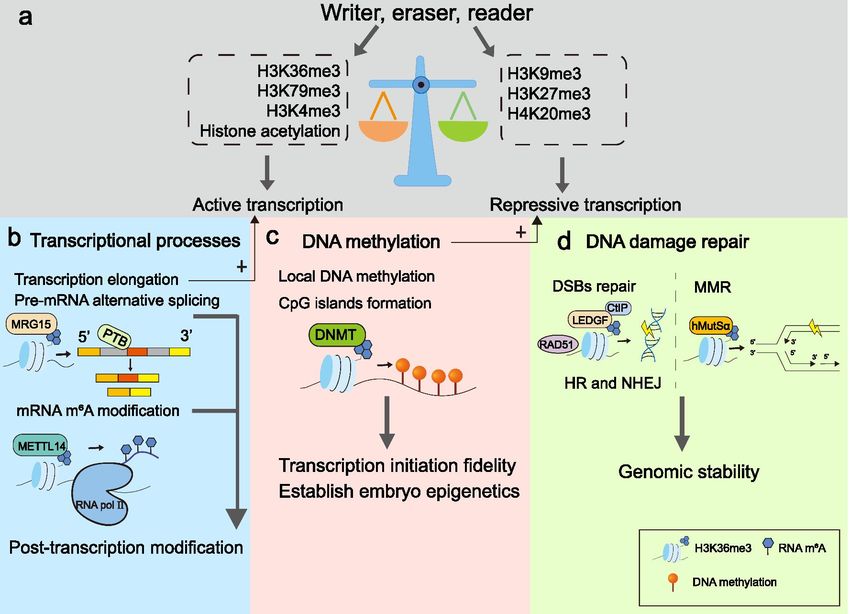 Fig.2 H3K36me3 engages in essential cellular possesses. (Xiao C, et al., 2021)
Fig.2 H3K36me3 engages in essential cellular possesses. (Xiao C, et al., 2021)Research Progress of HMTs
HMTs play a crucial role in the regulation of gene expression and chromatin structure by adding methyl groups to histone proteins. Recent research has made significant progress in understanding the functions and mechanisms of HMTs, as well as the application of CRISPR/Cas9 technology in studying histone methyltransferases.
| Types | Details |
|---|---|
| Function and Mechanisms of HMTs |
|
| CRISPR/Cas9 Technology in HMT Research |
|
| Emerging Technologies |
|
Overall, the research progress in histone methyltransferases has deepened our understanding of their biological functions and regulatory mechanisms. The integration of CRISPR/Cas9 technology and other advanced techniques has further accelerated discoveries in this field, opening up new avenues for therapeutic interventions and precision epigenome engineering.
Case Study
Case 1: Chen K, Liu J, Liu S, et al. Methyltransferase SETD2-mediated methylation of STAT1 is critical for interferon antiviral activity. Cell. 2017;170(3):492-506.e14.
Interferon-α (IFNα) signaling is crucial for antiviral response by inducing IFN-stimulated genes (ISGs). A study conducted high-throughput RNAi screening to identify epigenetic modifiers involved in IFNα-mediated inhibition of HBV replication. They discovered that methyltransferase SETD2 plays a critical role in enhancing IFNα-mediated antiviral immunity. Mice with hepatocyte-specific deletion of Setd2 showed increased susceptibility to HBV infection. SETD2 facilitates IFNα signaling by methylating STAT1 at lysine 525, enhancing STAT1 phosphorylation and antiviral response. Additionally, SETD2 selectively catalyzes tri-methylation of H3K36 on promoters of specific ISGs, such as ISG15, to activate gene expression.
The study highlights the essential role of SETD2-mediated STAT1 methylation and H3K36me3 modification in IFNα-dependent antiviral immunity and suggests SETD2 as a potential target for controlling viral infections.
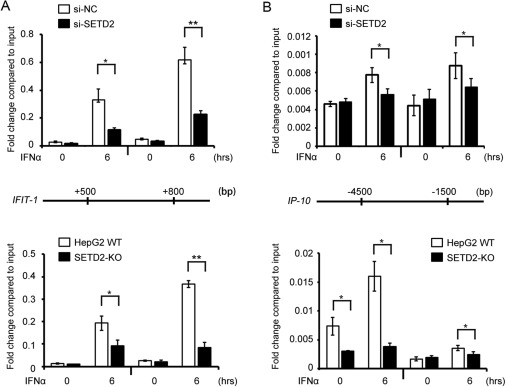 Fig.1 SETD2 selectively catalyzes H3K36me3 modification of a set of ISGs gene loci.
Fig.1 SETD2 selectively catalyzes H3K36me3 modification of a set of ISGs gene loci.Case 2: Ropa J, Saha N, Hu H, Peterson LF, Talpaz M, Muntean AG. SETDB1 mediated histone H3 lysine 9 methylation suppresses MLL-fusion target expression and leukemic transformation. Haematologica. 2020;105(9):2273-2285.
The study investigated the effects of SETDB1 expression on the transformation and growth of acute myeloid leukemia (AML) cells. They found that Setdb1 expression is reduced in mouse AML compared to normal hematopoietic stem and progenitor cells (HSPCs). Overexpression of human SETDB1 in AML cells significantly reduced colony formation and proliferation driven by the MLL-AF9 fusion oncogene and E2A-HLF fusion protein. SETDB1 overexpression also induced morphological changes consistent with differentiation in AML cells. Genes associated with hematopoietic differentiation showed increased expression upon SETDB1 overexpression. Additionally, SETDB1 overexpression led to increased apoptosis in AML cells. The study also examined the chemoresistance of AML cells and found that SETDB1 expression did not affect sensitivity to Daunorubicin treatment. Furthermore, overexpression of another H3K9 methyltransferase, G9A, had similar effects on AML cell proliferation, colony formation, and differentiation. These findings suggest that the expression of multiple H3K9 methyltransferases, including SETDB1 and G9A, can reduce AML cell growth, induce differentiation, and potentially serve as therapeutic targets for AML treatment.
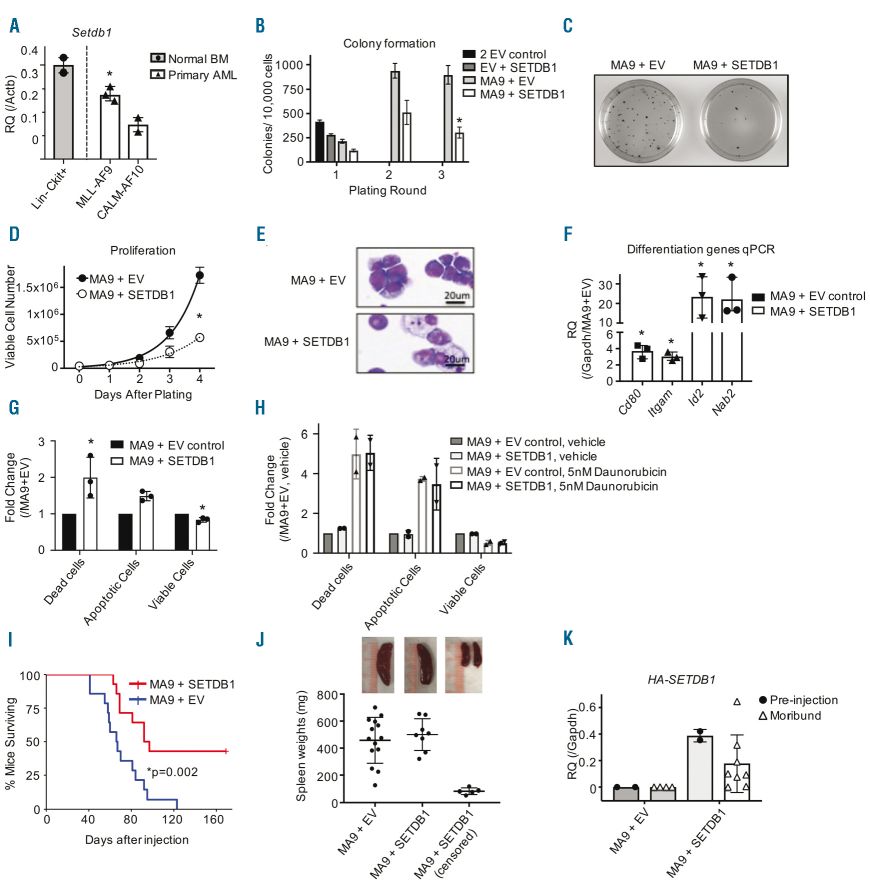 Fig.2 Overexpression of SETDB1 delays acute myeloid leukemia growth.
Fig.2 Overexpression of SETDB1 delays acute myeloid leukemia growth.Related References
- Chen Y, Liu X, Li Y, Quan C, Zheng L, Huang K. Lung Cancer therapy targeting histone methylation: opportunities and challenges. Comput Struct Biotechnol J. 2018;16:211-223.
- Xiao C, Fan T, Tian H, et al. H3K36 trimethylation-mediated biological functions in cancer. Clinical Epigenetics, 2021, 13(1):199-.
