DECR1
-
Official Full Name
2,4-dienoyl CoA reductase 1, mitochondrial -
Overview
This gene encodes an accessory enzyme which participates in the beta-oxidation and metabolism of unsaturated fatty enoyl-CoA esters. -
Synonyms
DECR1;2,4-dienoyl CoA reductase 1, mitochondrial;DECR;2,4-dienoyl-CoA reductase, mitochondrial;SDR18C1;short chain dehydrogenase/reductase family 18C;member 1;4-enoyl-CoA reductase;short chain dehydrogenase/reductase family 18C, member 1;NADPH
Recombinant Proteins
- Human
- Cynomolgus
- Zebrafish
- Mouse
- E.coli
- HEK293
- Mammalian Cell
- Wheat Germ
- HEK293T
- His
- Myc&DDK
- GST
- Non
- His&Fc&Avi
Background
What is DECR1 protein?
DECR1 gene (2,4-dienoyl-CoA reductase 1) is a protein coding gene which situated on the short arm of chromosome 8 at locus 8q21. DECR1 is primarily localized in mitochondria, where it is involved in metabolic processes and is part of the catalytic complex. DECR1 is A co-enzyme in the β-oxidation process, involved in the metabolism of unsaturated fatty acid acyl-coA ester. It catalyzes the NADP-dependent reduction of 2, 4-dienoyl-CoA to form trans-3-enoyl-CoA. The DECR1 protein is consisted of 1456 amino acids and DECR1 molecular weight is approximately 166.0 kDa.
What is the function of DECR1 protein?
DECR1 catalyzes a specific step in the beta-oxidation of unsaturated fatty acids. It reduces 2,4-dienoyl-CoA to trans-3-enoyl-CoA, a reaction that is NADP-dependent. It plays a crucial role in the metabolism of unsaturated fatty acids with double bonds in both even- and odd-numbered positions. Through its involvement in fatty acid metabolism, DECR1 contributes to the production of energy within cells by facilitating the breakdown of fatty acids. As a mitochondrial enzyme, DECR1 is part of the cellular respiration process that occurs within the mitochondria. Given its role in disease processes, DECR1 is considered a potential target for therapeutic intervention in conditions where its activity is dysregulated.
DECR1 Related Signaling Pathway
DECR1 is a key enzyme in the fatty acid beta-oxidation process, specifically acting as a rate-limiting enzyme in the metabolism of polyunsaturated fatty acids (PUFA). It is involved in converting PUFA into energy substances that can enter the mitochondria for oxidation, providing energy to the cell. DECR1 has been identified as a target gene for androgen receptors and is negatively regulated by androgens. The downregulation of DECR1 leads to the accumulation of PUFA, enhances mitochondrial oxidative stress and lipid peroxidation, and ultimately induces iron death, a form of cell death associated with oxidative stress. DECR1 may affect the survival and proliferation of tumor cells by regulating fatty acid metabolism in the tumor microenvironment, thus affecting tumor development and metastasis.
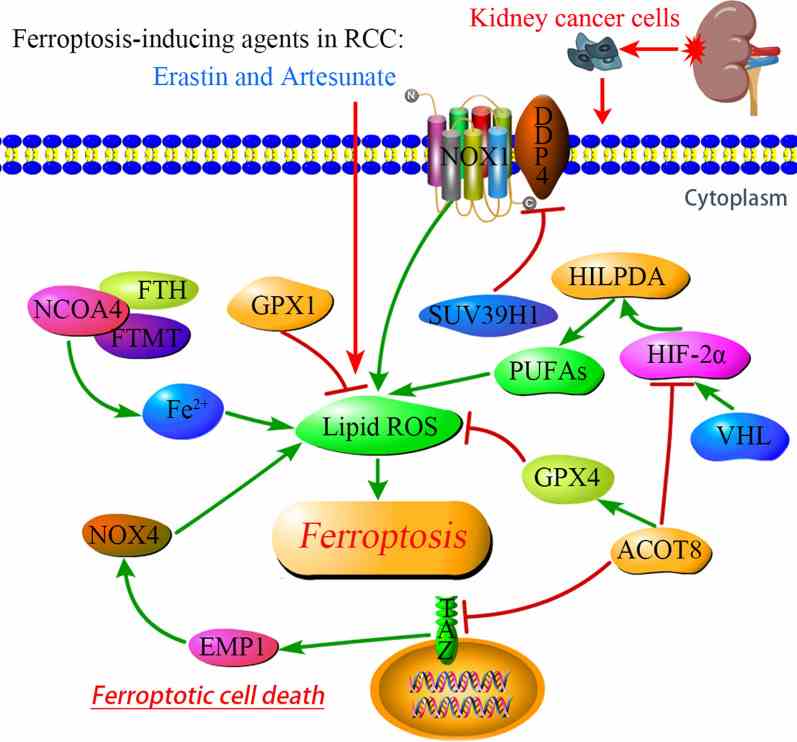
Fig1. Signaling pathways regulating ferroptosis in RCC. (Shankun Zhao, 2021)
DECR1 Related Diseases
DECR1 is abnormally high expressed in a variety of cancer types, including prostate cancer, breast cancer, and lung cancer, and its high expression is closely related to tumor aggressiveness, metastasis potential, and poor prognosis. In prostate cancer, DECR1 expression is negatively regulated by androgen receptor (AR) and is associated with treatment resistance. Down-regulation of DECR1 can inhibit the proliferation and migration of tumor cells, and induce iron death by enhancing mitochondrial oxidative stress and lipid peroxidation, thereby inhibiting tumor cell survival. In addition, DECR1 may also be involved in regulating other cellular processes such as apoptosis and autophagy.
Bioapplications of DECR1
The high expression of DECR1 in a variety of cancers suggests it could be a potential target for cancer therapy. Drug development targeting DECR1 may help inhibit the proliferation and metastasis of tumor cells. In certain inherited metabolic diseases, DECR1 activity tests may be useful for diagnosis and typing, such as those associated with abnormal fatty acid metabolism. As a key regulatory point of fatty acid metabolism, the functional study of DECR1 is helpful to further understand the pathogenesis of metabolic diseases such as obesity and diabetes. Personalized treatment design based on patient DECR1 expression levels has the potential to improve treatment outcomes and reduce unwanted side effects.
Case Study
Case Study 1: Zeyad D Nassar, 2020
Fatty acid β-oxidation (FAO) is the main bioenergetic pathway in human prostate cancer (PCa) and a promising novel therapeutic vulnerability. Here researchers demonstrate therapeutic efficacy of targeting FAO in clinical prostate tumors cultured ex vivo, and identify DECR1, encoding the rate-limiting enzyme for oxidation of polyunsaturated fatty acids (PUFAs), as robustly overexpressed in PCa tissues and associated with shorter relapse-free survival. DECR1 is a negatively-regulated androgen receptor (AR) target gene and, therefore, may promote PCa cell survival and resistance to AR targeting therapeutics. Mechanistically, targeting of DECR1 caused cellular accumulation of PUFAs, enhanced mitochondrial oxidative stress and lipid peroxidation, and induced ferroptosis.
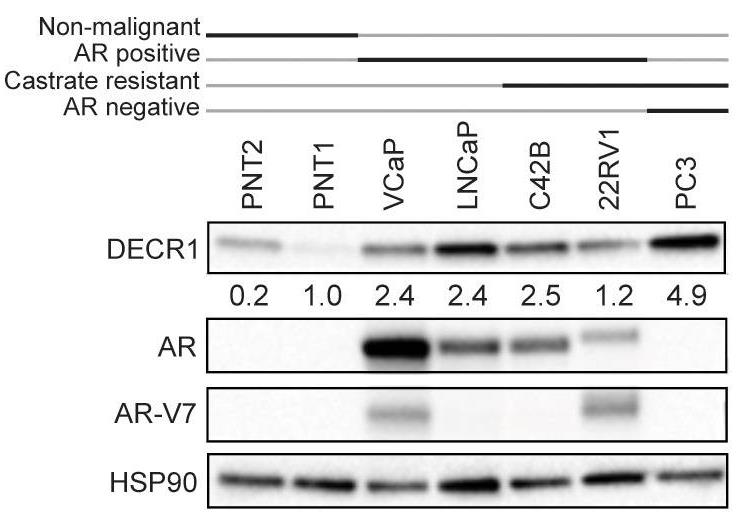
Fig1. DECR1 protein expression in non-malignant prostate cell lines and PCa cell lines.
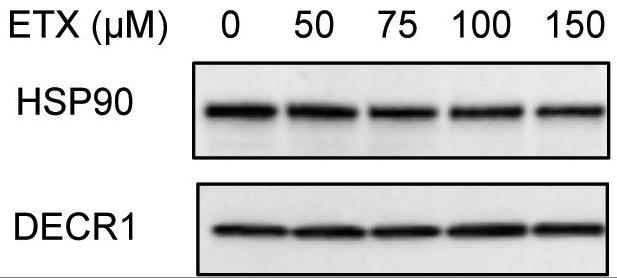
Fig2. DECR1 protein expression after 48 hr treatment with varying concentrations of etomoxir.
Case Study 2: Suiqun Guo, 2023
Immunotherapy has significantly improved patient outcomes, and genes related to tumor immune infiltration have been clinically relevant and highly reproducible biomarkers that affect the prognosis and response to treatment of CC. DECR1 was considered to be an oncogene in a previous study, but relationship between DECR1 and immune infiltration was not mentioned. This study aimed to reveal the clinical value of DECR1 in CC and to investigate its relationship with immune infiltration. Human Protein Atlas was used to identify the localization of DECR1. GSEA was used to assess the possible signaling pathways of DECR1 in CC. The TIMER database was applied to reveal the relevance between DECR1 and immune infiltration. GEPIA was conducted to detect the co-relationship among DECR1, immune markers, and typical molecules of apoptosis. The results showed that DECR1 was mainly distributed in the cytoplasm and overlapped with the endoplasmic reticulum. DECR1 was downregulated in CC compared to adjacent tissue. GSEA suggested that DECR1 was closely related to apoptosis signaling.
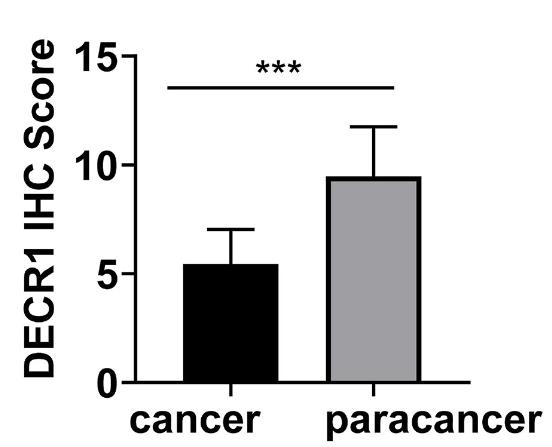
Fig3. Box plots assessing DECR1 expression between cervical cancer and adjacent samples.
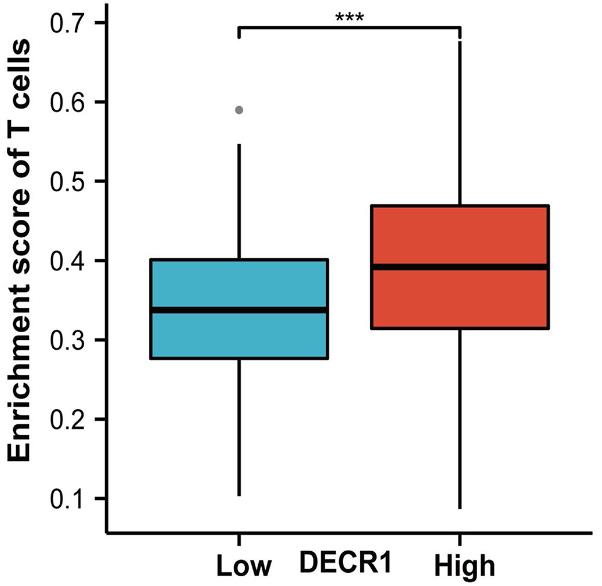
Fig4. Enrichment score of T cell in DECR1 high and low expression group.
Quality Guarantee
.
.jpg)
Fig1. SDS-PAGE (DECR1-26H)
Involved Pathway
DECR1 involved in several pathways and played different roles in them. We selected most pathways DECR1 participated on our site, such as Fatty Acid Beta Oxidation,Fatty Acid Biosynthesis,Fatty acid, triacylglycerol, and ketone body metabolism, which may be useful for your reference. Also, other proteins which involved in the same pathway with DECR1 were listed below. Creative BioMart supplied nearly all the proteins listed, you can search them on our site.
| Pathway Name | Pathway Related Protein |
|---|---|
| Fatty Acid Biosynthesis | ACSBG1,OLAH,ACSL3B,ACACA,ECH1,FASN,ACAA2,ACSL3A,OXSM,ACSL1 |
| Mitochondrial Fatty Acid Beta-Oxidation | ECI1,ACAD11 |
| Metabolism | GDF1,BRP44,HS3ST4,PFKLB,GRX2,CKMT2A,CTRC,ELOVL1A,CES1,SLC44A5B |
| Fatty acid, triacylglycerol, and ketone body metabolism | STIL,ACOT10,MED8,ACOT9.1,ACOT6,CYP7A1,HELZ2,ACOT11B,MTF1,ACOT9 |
| Fatty Acid Beta Oxidation | CPT1B,ECI1,CPT2,SLC25A20,CPT1A,CHKB,CRAT |
| Metabolism of lipids and lipoproteins | COQ6,ACER1,CRAT,ARSE,MED6,MED19,GLTPD1,CPNE6,CYP2AD6,SGMS2 |
Protein Function
DECR1 has several biochemical functions, for example, 2,4-dienoyl-CoA reductase (NADPH) activity,NADPH binding,oxidoreductase activity, acting on NAD(P)H. Some of the functions are cooperated with other proteins, some of the functions could acted by DECR1 itself. We selected most functions DECR1 had, and list some proteins which have the same functions with DECR1. You can find most of the proteins on our site.
| Function | Related Protein |
|---|---|
| oxidoreductase activity, acting on NAD(P)H | NDUFS8A,NDUFS8B,WDR93,DERL3,MIOX,AIFM1,ECSIT |
| NADPH binding | CBR3,CRYZ,TP53I3,QDPR,MTRR,CBR4,FASN,KCNAB1,HMGCR,GRHPR |
| 2,4-dienoyl-CoA reductase (NADPH) activity | DECR2 |
Interacting Protein
DECR1 has direct interactions with proteins and molecules. Those interactions were detected by several methods such as yeast two hybrid, co-IP, pull-down and so on. We selected proteins and molecules interacted with DECR1 here. Most of them are supplied by our site. Hope this information will be useful for your research of DECR1.
PTTG1;thiI;ssrna_au;Ktn1;NOP56;CCDC22;AK1;ESR1;NPM1;VCAM1;NSS;RPL10;JAK3;MSH6
Resources
Related Services
Related Products
References
- Filppula, SA; Yagi, AI; et al. Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase from rat liver - Molecular characterization. JOURNAL OF BIOLOGICAL CHEMISTRY 273:349-355(1998).
- KajiharaKano, H; Hayakari, M; et al. Characterization of S-hexylglutathione-binding proteins of human hepatocellular carcinoma: separation of enoyl-CoA isomerase from an Alpha class glutathione transferase form. BIOCHEMICAL JOURNAL 328:473-478(1997).


