MSH6
-
Official Full Name
mutS homolog 6 -
Overview
MSH6 is a mismatch repair gene which is deficient in a high proportion of patients with microsatellite instability (MSI-H). This finding is associated with the autosomal dominant condition known as Hereditary Non-Polyposis Colon Cancer (HNPCC). The anti-M -
Synonyms
MSH6;mutS homolog 6 (E. coli);GTBP, mutS (E. coli) homolog 6;DNA mismatch repair protein Msh6;p160;GTMBP;hMSH6;sperm-associated protein;mutS-alpha 160 kDa subunit;G/T mismatch-binding protein;GTBP;HSAP;HNPCC5
Recombinant Proteins
- Mouse
- Human
- Rhesus macaque
- Zebrafish
- Mammalian Cells
- E.coli
- Wheat Germ
- HEK293
- His
- GST
- Non
- Avi
- Fc
- SUMO
Background
What is MSH6 protein?
MSH6 (mutS homolog 6) gene is a protein coding gene which situated on the short arm of chromosome 2 at locus 2p16. MSH6 is a crucial component of the DNA mismatch repair (MMR) system in eukaryotic cells. MSH6 is encoded by the MSH6 gene and functions by forming a heterodimer with MSH2, which is referred to as MutS-alpha complex. This complex is instrumental in recognizing and repairing errors that occur during DNA replication, such as single-base pair mismatches and small insertions or deletions of nucleotides. The assembly of the MutS-alpha complex is the initial step in the MMR process, which is vital for maintaining genomic stability and preventing mutations that can lead to various diseases, including cancer. The MSH6 protein is consisted of 1360 amino acids and its molecular mass is approximately 152.8 kDa.
What is the function of MSH6 protein?
MSH6 protein, a key component of the DNA mismatch repair (MMR) system in eukaryotes, plays an essential role in maintaining genomic stability and preventing mutations that can lead to cancer and other diseases. The biological function of MSH6 is not limited to the MMR pathway. It has also been implicated in other cellular processes, including meiosis, where it contributes to the proper segregation of chromosomes and the prevention of aneuploidy, a condition where cells have an abnormal number of chromosomes. Furthermore, MSH6 interacts with other proteins, such as PCNA (Proliferating Cell Nuclear Antigen), which is involved in DNA replication and repair, suggesting that MSH6 may have additional roles in these processes. MSH6's role in DNA repair is underscored by its association with Lynch syndrome, an inherited condition that increases the risk of developing certain types of cancers, particularly colorectal cancer.
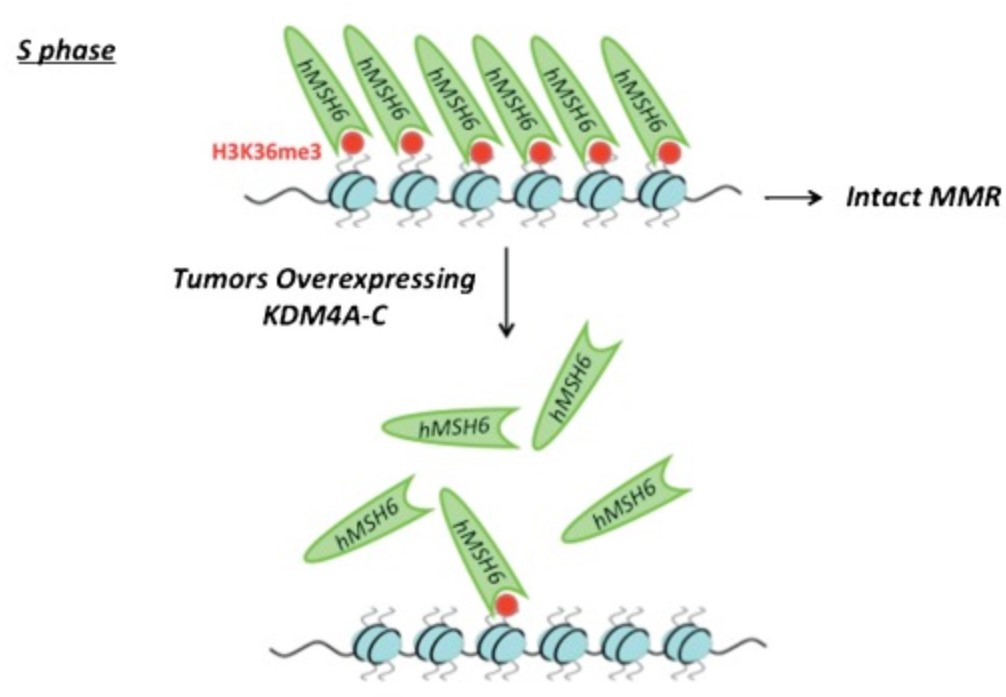
Fig1. A model showing that KDM4A-C overexpression diminishes H3K36me3 signal, impairs MSH6 foci and impairs the integrity of DNA MMR pathway. (Samah W Awwad, 2015)
MSH6 Related Signaling Pathway
MSH6 forms heterodimers with MSH2, recognizes and binds to base mismatch sites on DNA, and then interacts with other MMR proteins (such as MLH1, PMS2, etc.) to initiate the mismatch repair process. This signaling pathway plays a key role in maintaining genomic stability and preventing mutation accumulation. The DNA mismatch repair process involved in MSH6 is closely related to cell cycle regulation. The DNA mismatch repair process involved in MSH6 is also a cellular response mechanism to DNA damage. The DNA mismatch repair process involved in MSH6 may affect the transcriptional activity of genes.
MSH6 Related Diseases
MSH6 protein is associated with several diseases, particularly those related to the DNA mismatch repair (MMR) system. The most notable disease associated with MSH6 is hereditary non-polyposis colon cancer (HNPCC), also known as Lynch syndrome. This condition is characterized by an increased risk of developing colorectal cancer and other types of cancers, such as endometrial and uterine cancers. Mutations in the MSH6 gene can lead to a dysfunctional MMR system, resulting in microsatellite instability (MSI) and an increased mutation rate, which is a hallmark of many cancers.
In addition to its role in Lynch syndrome, MSH6 has also been implicated in other cancers. Furthermore, MSH6 has been linked to other conditions such as type 1 neurofibromatosis, neutropenia, and various other syndromes and tumors.
Bioapplications of MSH6
MSH6, in conjunction with other MMR proteins, is used as a biomarker to identify cancers with microsatellite instability (MSI). This is particularly relevant in colorectal cancer, endometrial cancer, and other tumor types. Given its role in DNA repair, MSH6 is considered a potential therapeutic target for cancer treatment. The study of MSH6 and its interactions with other proteins and DNA can inform the design of molecular diagnostic tools.
Case Study
Case Study 1: Yu Zong, 2024
Arginine and lysine, frequently appearing as a pair on histones, have been proven to carry diverse modifications and execute various epigenetic regulatory functions. However, the most context-specific and transient effectors of these marks, while significant, have evaded study as detection methods have thus far not reached a standard to capture these ephemeral events. Herein, a pair of complementary photo-arginine/δ-photo-lysine (R-dz/K-dz) probes is developed and involve these into histone peptide, nucleosome, and chromatin substrates to capture and explore the interactomes of Arg and Lys hPTMs. By means of these developed tools, this study identifies that H3R2me2a can recruit MutS protein homolog 6 (MSH6), otherwise repelDouble PHD fingers 2 (DPF2), Retinoblastoma binding protein 4/7 (RBBP4/7). And it is disclosed that H3R2me2a inhibits the chromatin remodeling activity of the cBAF complex by blocking the interaction between DPF2 (one component of cBAF) and the nucleosome. In addition, the novel pairs of H4K5 PTMs and respective readers are highlighted, namely H4K5me-Lethal(3)malignant brain tumor-like protein 2 (L3MBTL2), H4K5me2-L3MBTL2, and H4K5acK8ac-YEATS domain-containing protein 4 (YEATS4).
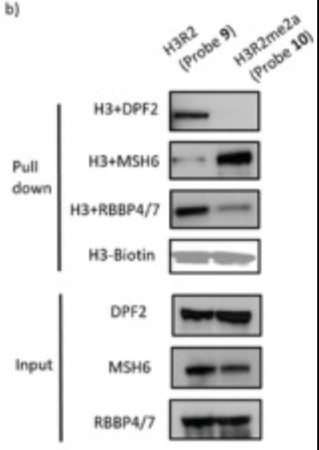
Fig1. Western blot analysis of probe 9 and probe 10 pull-down interacting protein.
-H3R2me2a (probe 12).jpg)
Fig2. Western blot analysis of interacting protein enriched by H3R2 (probe 11)/H3R2me2a (probe 12).
Case Study 2: Lin Wang, 2023
DNA mismatch repair (MMR) improves replication accuracy by up to three orders of magnitude. The MutS protein in E. coli or its eukaryotic homolog, the MutSα (Msh2-Msh6) complex, recognizes base mismatches and initiates the mismatch repair mechanism. Msh6 is an essential protein for assembling the heterodimeric complex. However, the function of the Msh6 subunit remains elusive. Tetrahymena undergoes multiple DNA replication and nuclear division processes, including mitosis, amitosis, and meiosis. Here, Msh6Tt localized in the macronucleus (MAC) and the micronucleus (MIC) during the vegetative growth stage and starvation. During the conjugation stage, Msh6Tt only localized in MICs and newly developing MACs. MSH6Tt knockout led to aberrant nuclear division during vegetative growth. The MSH6TtKO mutants were resistant to treatment with the DNA alkylating agent methyl methanesulfonate (MMS) compared to wild type cells. MSH6Tt knockout affected micronuclear meiosis and gametogenesis during the conjugation stage. Furthermore, Msh6Tt interacted with Msh2Tt and MMR-independent factors. Downregulation of MSH2Tt expression affected the stability of Msh6Tt. In addition, MSH6Tt knockout led to the upregulated expression of several MSH6Tt homologs at different developmental stages.
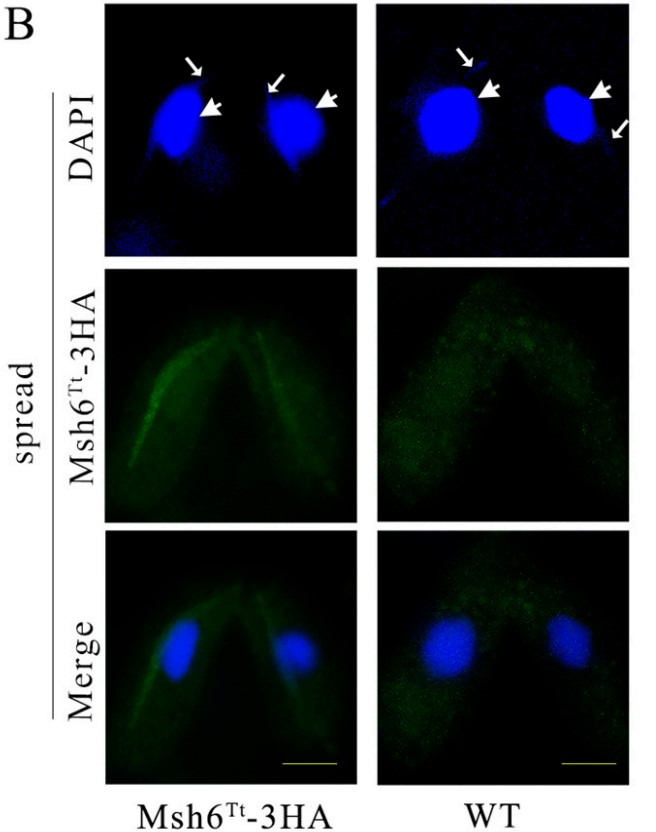
Fig3. Msh6Tt-3HA localized on chromatin in spread cells.
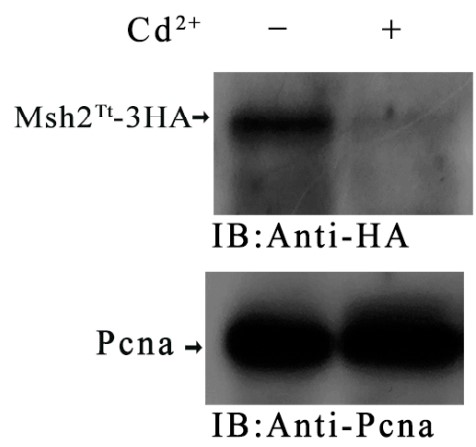
Fig4. The expression level of Msh2Tt-3HA and Msh6Tt-3HA when the msh2i mutants mating with Msh2Tt-3HA or Msh6Tt-3HA mutants.
Quality Guarantee
High Purity
.jpg)
Fig1. SDS-PAGE (MSH6-4225H)
.
.jpg)
Fig2. SDS-PAGE (MSH6-03H)
Involved Pathway
MSH6 involved in several pathways and played different roles in them. We selected most pathways MSH6 participated on our site, such as Mismatch repair,Pathways in cancer,Colorectal cancer, which may be useful for your reference. Also, other proteins which involved in the same pathway with MSH6 were listed below. Creative BioMart supplied nearly all the proteins listed, you can search them on our site.
| Pathway Name | Pathway Related Protein |
|---|---|
| Mismatch repair | PCNA,RPA4,RPA2,RFC5,SSBP1,RFC4,EXO1,MSH3,PMS2,MLH3 |
| Pathways in cancer | WNT7A,PLCB2,MAP2K2,BID,WNT5B,TRAF5,BRAF,RHOA,VEGFA,WNT2B |
| Colorectal cancer | CYCS,BAD,CCND1,SMAD2,ARAF,MSH2,TGFBR2,PIK3R1,TRP53,GSK3B |
Protein Function
MSH6 has several biochemical functions, for example, contributes_to ADP binding,contributes_to ATP binding,contributes_to ATPase activity. Some of the functions are cooperated with other proteins, some of the functions could acted by MSH6 itself. We selected most functions MSH6 had, and list some proteins which have the same functions with MSH6. You can find most of the proteins on our site.
| Function | Related Protein |
|---|---|
| contributes_to ATP binding | ATP1B1,MSH2,SFRP5,ATP1A2,ATP5D,ABCG8 |
| contributes_to single thymine insertion binding | MSH2 |
| contributes_to MutLalpha complex binding | MSH2 |
| protein binding | USP6,F8,IL1RN,ATRNL1,ZNF593,ETV6,DLA,ALCAM,SNF8,PPP3CA |
| DNA-dependent ATPase activity | IGHMBP2,MSH2,RAD51B,RAD51L1,RAD51C,RAD51L3,TTF2,CHD8,RAD51D,DDX11 |
| NOT protein homodimerization activity | MLXIPL,DDIT3,MSH3,MYO9B,EXT2,NUDT16L1,MECOM,KCNA3 |
| contributes_to mismatched DNA binding | MSH3,MSH2 |
| contributes_to ATPase activity | RFC3,ATP1B1,ATP5A1,ATP5D,ATP5B,ATP5E,ATP5C1,PPP2R4,ATP5F1,ABCG8 |
| contributes_to magnesium ion binding | MSH2 |
Interacting Protein
MSH6 has direct interactions with proteins and molecules. Those interactions were detected by several methods such as yeast two hybrid, co-IP, pull-down and so on. We selected proteins and molecules interacted with MSH6 here. Most of them are supplied by our site. Hope this information will be useful for your research of MSH6.
MSH2;MYC;41_bp_heteroduplex;BRCA1;MCC;CASP4;PSMD4;HRAS;MAP2K1;q5ngv3_fratt;NANS;midostaurin
Resources
Related Services
Related Products
References
- Stark, AM; Doukas, A; et al. Expression of DNA mismatch repair proteins MLH1, MSH2, and MSH6 in recurrent glioblastoma. NEUROLOGICAL RESEARCH 37:95-105(2015).
- Jentzsch, T; Robl, B; et al. Expression of MSH2 and MSH6 on a Tissue Microarray in Patients with Osteosarcoma. ANTICANCER RESEARCH 34:6961-6972(2014).


