HDAC5
-
Official Full Name
histone deacetylase 5 -
Overview
Histones play a critical role in transcriptional regulation, cell cycle progression, and developmental events. Histone acetylation/deacetylation alters chromosome structure and affects transcription factor access to DNA. The protein encoded by this gene belongs to the class II histone deacetylase/acuc/apha family. It possesses histone deacetylase activity and represses transcription when tethered to a promoter. It coimmunoprecipitates only with HDAC3 family member and might form multicomplex proteins. It also interacts with myocyte enhancer factor-2 (MEF2) proteins, resulting in repression of MEF2-dependent genes. This gene is thought to be associated with colon cancer. Two transcript variants encoding different isoforms have been found for this gene. -
Synonyms
HDAC5;histone deacetylase 5;FLJ90614;KIAA0600;NY CO 9;antigen NY-CO-9;HD5;NY-CO-9
Recombinant Proteins
- Human
- Mouse
- Rhesus macaque
- Insect Cell
- Sf9 Insect Cell
- HEK293 cells
- Mammalian Cell
- Wheat Germ
- HEK293
- Insect cell
- E.coli
- GST
- His
- Non
- His&Fc&Avi
Background
What is HDAC5 protein?
HDAC5 gene (histone deacetylase 5) is a protein coding gene which situated on the long arm of chromosome 17 at locus 17q21. Histones play a critical role in transcriptional regulation, cell cycle progression, and developmental events. Histone acetylation/deacetylation alters chromosome structure and affects transcription factor access to DNA. The protein encoded by this gene belongs to the class II histone deacetylase/acuc/apha family. It possesses histone deacetylase activity and represses transcription when tethered to a promoter. It coimmunoprecipitates only with HDAC3 family member and might form multicomplex proteins. It also interacts with myocyte enhancer factor-2 (MEF2) proteins, resulting in repression of MEF2-dependent genes. This gene is thought to be associated with colon cancer. The HDAC5 protein is consisted of 1122 amino acids and HDAC5 molecular weight is approximately 122.0 kDa.
What is the function of HDAC5 protein?
It plays a crucial role in the regulation of gene expression and is known to undergo nuclear-cytoplasmic shuttling, acting as a critical transcriptional regulator. HDAC5 is involved in various cellular processes, including cardiac development, neuronal function, and cell cycle regulation. Its activity can be modulated by post-translational modifications such as phosphorylation, which can affect its subcellular localization, interactions with other proteins, and enzymatic activity. HDAC5's function can be studied through various experimental methods that assess its subcellular localization, protein interactions, post-translational modifications, and activity.
HDAC5 Related Signaling Pathway
HDAC5 plays a role in the vascular endothelial growth factor (VEGF) signaling pathway and influences VEGF-mediated angiogenesis by binding to myocyte enhancer factor 2 (MEF2). The expression of HDAC5 is regulated by the PI3K/Akt signaling pathway, which influences the activity and expression of HDAC5 through changes in m6A RNA methylation levels, thereby affecting the epithelial-mesenchymal transformation (EMT) of renal tubular cells in diabetic nephropathy. HDAC5 may be involved in the regulation of endothelial angiogenesis through Notch signaling pathway. HDAC5 interacts with the tumor suppressor protein p53, influencing p53 target selection and function, including promoting cell cycle arrest and antioxidant effects.
HDAC5 Related Diseases
HDAC5 is implicated in a variety of diseases, particularly those related to cardiac and neurological functions, as well as in the development of cancer. It is known to play a role in cardiac hypertrophy, heart failure, and vascular diseases, where its regulation of gene expression can influence cardiac cell growth and function. In the context of neurological disorders, HDAC5 has been associated with stroke and cerebral ischemia, potentially affecting vascularization and neuronal survival post-ischemic events. Moreover, HDAC5 has been linked to cancer development and progression, with its misregulation potentially leading to uncontrolled cell proliferation and tumor growth. Additionally, HDAC5's involvement in diabetes has been noted, suggesting its role in metabolic regulation.

Fig1. Proposed model of the role of HDAC5-LSD1 axis in breast cancer development. (C Cao, 2017)
Bioapplications of HDAC5
HDAC5 is considered a credible molecular target for anticancer therapeutics due to its significant variation in expression between cancer patients and healthy individuals. HDAC5 has been implicated in the development and progression of breast cancer, with its structure and function being the subject of intensive research for the design of HDAC5 inhibitors. Beyond cancer, HDAC5's bioapplications extend to the study of its role in muscle maturation and the repression of myocyte enhancer MEF2C, with its activity being modulated during muscle differentiation.
Case Study
Case Study 1: C Cao, 2017
Researchers have previously demonstrated that crosstalk between lysine-specific demethylase 1 (LSD1) and histone deacetylases (HDACs) facilitates breast cancer proliferation. However, the underlying mechanisms are largely unknown. Here, they report that expression of HDAC5 and LSD1 proteins were positively correlated in human breast cancer cell lines and tissue specimens of primary breast tumors. Protein expression of HDAC5 and LSD1 was significantly increased in primary breast cancer specimens in comparison with matched-normal adjacent tissues. Using HDAC5 deletion mutants and co-immunoprecipitation studies, they showed that HDAC5 physically interacted with the LSD1 complex through its domain containing nuclear localization sequence and phosphorylation sites. The in vitro acetylation assays revealed that HDAC5 decreased LSD1 protein acetylation. Overexpression of HDAC5 stabilized LSD1 protein and decreased the nuclear level of H3K4me1/me2 in MDA-MB-231 cells. Researchers further demonstrated that HDAC5 promoted the protein stability of USP28, a bona fide deubiquitinase of LSD1.

Fig1. IB with anti-FLAG was used to detect the levels of FLAG-tagged HDAC5 full-length or deletion mutants.

Fig2. Effect of HDAC5 on protein acetylation of LSD1/USP28 and transcription of LSD1 target genes.
Case Study 2: Ming-Min Chang, 2024
Previous work demonstrated that chitosan nano-deposited surfaces induce spheroid formation and differentiation of ASCs for treating sciatic nerve injuries. However, the underlying cell fate and differentiation mechanisms of ASC-derived spheroids remain unknown. Here, researchers investigate the epigenetic regulation and signaling coordination of these therapeutic spheroids. During spheroid formation, they observed significant increases in histone deacetylase (HDAC) activities and decreased histone acetyltransferase activities. Additionally, HDAC5 translocated from the cytoplasm to the nucleus, along with increased nuclear HDAC5 activities. Utilizing single-cell RNA sequencing (scRNA-seq), they analyzed the chitosan-induced ASC spheroids and discovered distinct cluster subpopulations, cell fate trajectories, differentiation traits, and signaling networks using the 10x Genomics platform, R studio/language, and the Ingenuity Pathway Analysis (IPA) tool.
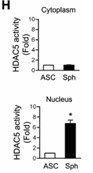
Fig3. HDAC5 activities in cytoplasm and nucleus of spheroids after 72 hours of chitosan induction.
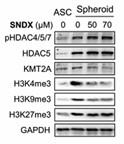
Fig4. Interactions between inhibitors of epigenetic regulators were shown in Western blotting analysis.
Quality Guarantee
Involved Pathway
HDAC5 involved in several pathways and played different roles in them. We selected most pathways HDAC5 participated on our site, such as Alcoholism,B Cell Receptor Signaling Pathway,Cell cycle, which may be useful for your reference. Also, other proteins which involved in the same pathway with HDAC5 were listed below. Creative BioMart supplied nearly all the proteins listed, you can search them on our site.
| Pathway Name | Pathway Related Protein |
|---|---|
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | ADAM10,HEYL,HEY2,JAG2,CCNC,HDAC6,HEY1,MIB1,CDK8,JAG1 |
| Diseases of signal transduction | STRA6,TTR,CCNC,RNF43,TNKS,DERL2,HEYL,FKBP1A,FGFR1OP,TRAT1 |
| Cell cycle | ORC2L,TFDP2,PIANP,TERF2IP,TERF2,MCM10,CDKN2A,RCC2,YWHABL,SUN1 |
| Alcoholism | SLC18A2,GNG10,HDAC3,HIST1H2BE,HIST1H2AI,GNAI3,H2AFB1,HIST1H3C,GRIN1,HIST1H2BF |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | HEY2,HEY1,ADAM10,JAG1,MIB1,HEYL,HDAC6,JAG2,CCNC,CDK8 |
| Disease | OS9,OPN1MW,TCEB3,AP1M2,FDX1L,NUPL2,KPNA5,OPN1SW,TIRAP,MIB1 |
| Epstein-Barr virus infection | TRAF3,HLA-DRB1,Fcer2a,LYN,IFNG,YWHAH,CSNK2B,HLA-DPA1,PIK3CG,HSPA1L |
| B Cell Receptor Signaling Pathway | AKT2,PIK3R1,PTPN18,NEDD9,RAP2A,PPP3CB,NFKBIB,DUSP6,PRKCB,PLEKHA2 |
Protein Function
HDAC5 has several biochemical functions, for example, NAD-dependent histone deacetylase activity (H3-K14 specific),RNA polymerase III transcription factor binding,chromatin binding. Some of the functions are cooperated with other proteins, some of the functions could acted by HDAC5 itself. We selected most functions HDAC5 had, and list some proteins which have the same functions with HDAC5. You can find most of the proteins on our site.
| Function | Related Protein |
|---|---|
| transcription factor binding | HMGA2,DRG1,RBL1,INSIG2,TNFRSF10A,TFDP1,TRERF1,SMAD2,PIAS2,JUNBA |
| protein deacetylase activity | HDAC2,SIRT2,HDAC1,HDAC10,SIRT1,HDAC4,SIN3A,HDAC3,HDAC9 |
| contributes_to transcription regulatory region DNA binding | TCF3,NFYB,ATF4,TAF1,TCF12,NR1H3,HAND1,HDAC4,RXRA,NFYA |
| histone deacetylase binding | SIRT2,GLI3,RELA,USF1,DNMT3B,MECP2,NKX2-5,PARK2,MAGEA2,SIX3 |
| protein binding | USP46,KIAA1199,PRDM1,NAT10,CXXC11,DPYSL4,KISS1,ASB3,MVK,TACC3 |
| NAD-dependent histone deacetylase activity (H3-K14 specific) | HDAC7,HDAC4,HDAC8,HDAC9,HDAC9B,HDAC6,HDAC10,HDAC7A,HDAC2,HDAC11 |
| histone deacetylase activity | SIRT1,HDAC9,HDAC8,NACC2,HDAC11,SIRT2,HDAC1,HDAC9B,HDAC10,HDAC3 |
| metal ion binding | PAPD4,ZNF131,GIT2A,ADPRHL2,FTR54,CYP2C37,KCNIP1B,ZBTB16A,NRP2A,RPS27A |
| chromatin binding | BRD4,TAL1,JUB,ARID3C,NCOA1,TPR,KDM5A,GATA2A,MSL1A,LDB1 |
Interacting Protein
HDAC5 has direct interactions with proteins and molecules. Those interactions were detected by several methods such as yeast two hybrid, co-IP, pull-down and so on. We selected proteins and molecules interacted with HDAC5 here. Most of them are supplied by our site. Hope this information will be useful for your research of HDAC5.
YWHAZ;SFN;YWHAB;YWHAE;ICP0;BRMS1;RUNX3
HDAC5 Related Signal Pathway
Resources
Related Services
Related Products
References
- Chang, CW; Cheong, ML; et al. Involvement of Epac1/Rap1/CaMKI/HDAC5 signaling cascade in the regulation of placental cell fusion. MOLECULAR HUMAN REPRODUCTION 19:745-755(2013).
- Maxwell, JT; Natesan, S; et al. Modulation of Inositol 1,4,5-Trisphosphate Receptor Type 2 Channel Activity by Ca2+/Calmodulin-dependent Protein Kinase II (CaMKII)-mediated Phosphorylation. JOURNAL OF BIOLOGICAL CHEMISTRY 287:-(2012).


.jpg)
.jpg)