MITF
-
Official Full Name
microphthalmia-associated transcription factor -
Overview
This gene encodes a transcription factor that contains both basic helix-loop-helix and leucine zipper structural features. It regulates the differentiation and development of melanocytes retinal pigment epithelium and is also responsible for pigment cell-specific transcription of the melanogenesis enzyme genes. Heterozygous mutations in the this gene cause auditory-pigmentary syndromes, such as Waardenburg syndrome type 2 and Tietz syndrome. Alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Jul 2008] -
Synonyms
MITF;microphthalmia-associated transcription factor;MI;WS2;CMM8;WS2A;bHLHe32;class E basic helix-loop-helix protein 32
Recombinant Proteins
- Human
- Rhesus macaque
- Chicken
- Mouse
- Aspergillus fumigatus
- Neosartorya fumigata
- E.coli
- Mammalian Cells
- HEK293
- Wheat Germ
- In Vitro Cell Free System
- His
- Non
- GST
- Avi
- Fc
- Myc
- SUMO
Background
What is MITF protein?
MITF gene (melanocyte inducing transcription factor) is a protein coding gene which situated on the short arm of chromosome 3 at locus 3p13. MITF is a transcription factor that plays a role in a variety of cell types and is particularly critical in the development and function of melanocytes. MITF regulates the expression of downstream genes by binding to M-box motifs on specific DNA sequences, which are typically involved in melanin production, cell proliferation, differentiation, and survival. MITF also plays a key role in the development of melanoma and is the ultimate target of many signal transduction pathways. The MITF protein is consisted of 526 amino acids and MITF molecular weight is approximately 58.8 kDa.
What is the function of MITF protein?
The MITF protein is a key transcription factor that plays a crucial role in the development and function of multiple cell types, particularly in melanocytes, which are responsible for the production of the pigment melanin. MITF regulates the production of melanin by regulating the expression of genes associated with melanin synthesis, including tyrosinase (TYR), TRP-1, and TRP-2, and binds specific DNA sequences in the promoter region of these genes. It is involved in the survival of melanocyte precursors, preventing apoptosis, and promoting the differentiation of melanocytes and other cell types. In melanocytes, MITF acts as a tumor suppressor and is often down-regulated in melanoma, suggesting that it plays a role in preventing the development of this skin cancer. In addition, post-translational modifications such as phosphorylation and acetylation can regulate MITF activity, DNA-binding affinity, and subcellular localization.
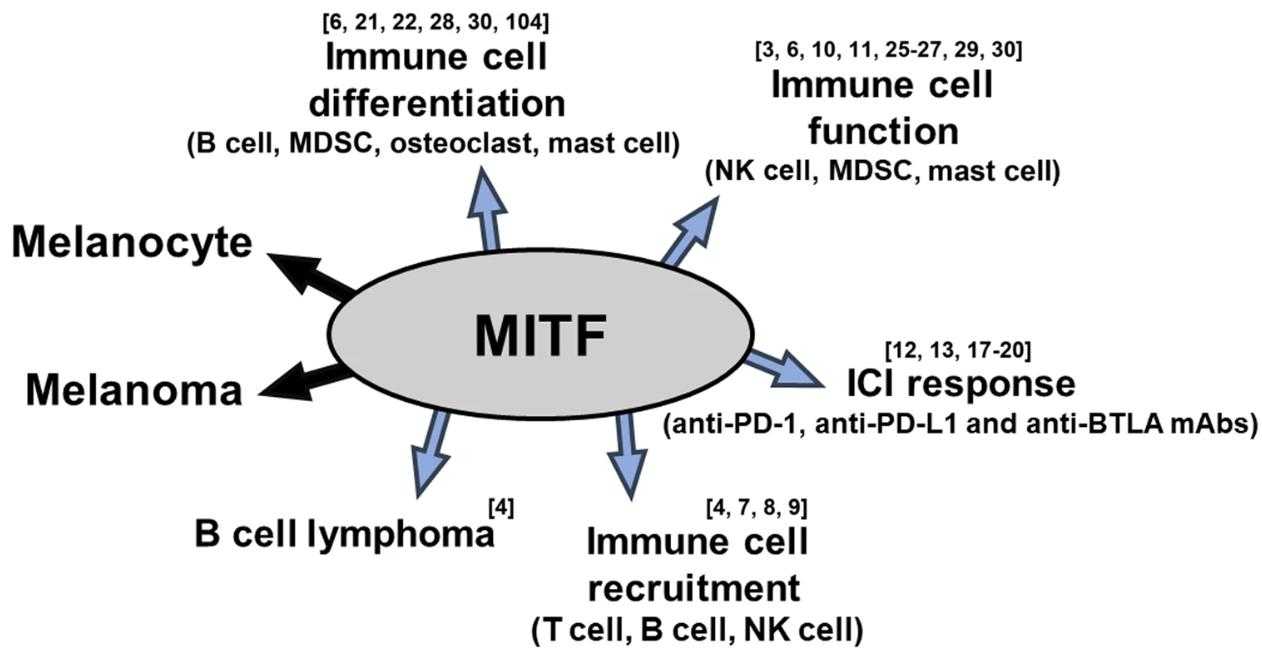
Fig1. The schematic shows the multiple roles of MITF. (Aram Lee, 2024)
MITF related signaling pathway
MITF protein is a key transcription factor that regulates melanin production and is involved in several signaling pathways. Among them, the cAMP/PKA signaling pathway is activated by binding to the MC1R receptor to increase cAMP concentration and further promote MITF expression. In the PI3K/Akt signaling pathway, GSK3β can be phosphorylated after Akt activation, which affects the activity and stability of MITF. MAPK signaling pathway promotes MITF expression and melanin production through the activation of ERK and p38. Wnt/β-catenin signaling pathway up-regulates MITF in melanocytes by influencing the accumulation of β-catenin in the nucleus. NO signaling pathway produces cGMP through soluble guanylate cyclase (sGC), activates PKG, and increases MITF expression. These pathways jointly regulate the development, proliferation, differentiation and migration of melanocytes and are essential for melanin production.
MITF related diseases
MITF protein is a transcription factor, and its abnormal function is associated with the occurrence and development of many diseases. Mutations or dysregulated expression of genes can lead to abnormal pigment production, such as Waardenburg syndrome, a genetic disorder that affects eye color and hearing. MITF plays a key role in the development of melanoma and other tumors, affecting the proliferation, invasion and drug resistance of tumor cells. In addition, MITF also participates in the differentiation and function of immune cells, regulates the activity of macrophages, dendritic cells and natural killer cells, and has an important impact on immune response. Abnormal expression of MITF is also associated with acute myeloid leukemia and other blood diseases, affecting the normal hematopoietic process. In addition to these, MITF may also be associated with a variety of pathological processes such as neurodegenerative diseases and cardiovascular diseases.
Bioapplications of MITF
MITF protein has a wide range of applications in the biomedical field, especially in the treatment and diagnosis of diseases show important value. In melanoma treatment, MITF, as a key transcriptional regulatory factor, affects the proliferation, invasion and drug resistance of tumor cells. Inhibitors targeting MITF are under development. In addition, MITF also plays a role in the immune response, and its expression changes may affect the function of macrophages, dendritic cells and natural killer cells. In blood diseases such as acute myeloid leukemia (AML), abnormal expression of MITF can affect the normal hematopoietic process. At the same time, the acetylation status of MITF changes its binding specificity to DNA and its distribution on the genome, which may have an important impact on the occurrence and development of related diseases. Therefore, MITF, as a potential biomarker or therapeutic target, is of great significance in the basic research and clinical application of diseases.
Case Study
Case Study 1: Pakavarin Louphrasitthiphol, 2023
Transcription factor MITF is vital for melanocyte development and melanoma, influencing processes like proliferation, differentiation, and invasion. The mechanism by which MITF selects between binding sites for these functions is unclear. This study reveals that MITF has an unusually long residence time on DNA, which is shortened by acetylation at K206 by p300/CBP. This modification reduces MITF's overall DNA-binding affinity but specifically redirects it from differentiation-associated CATGTG motifs to proliferation-linked CACGTG sites.
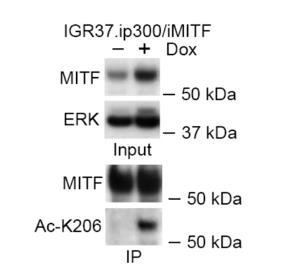
Fig1. Immunoprecipitation of HA-tagged MITF from IGR37 cells engineered to express doxycycline inducible p300 and HA-MITF.
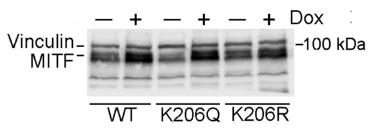
Fig2. Western blot of HALO-tagged MITF WT and mutants in 501mel cells with or without treatment with Doxycycline.
Case Study 2: Elizabeth Proaño-Pérez, 2023
MITF protein plays a significant role in a variety of diseases, primarily associated with pigmentation and tumor development. Mutations in the MITF gene can lead to pigmentation disorders such as Waardenburg syndrome, which affects eye color and hearing. In oncology, MITF functions as a critical regulator in melanoma, where its expression level influences tumor progression and response to treatment. High MITF expression is linked to poorer outcomes in certain cancers like diffuse large B-cell lymphoma. Furthermore, MITF is implicated in immune responses, affecting the function and recruitment of immune cells in diseases and cancer, and it may also play a role in the response to immune checkpoint inhibitors. Recent research has also highlighted MITF's association with immune cell differentiation and activity, influencing the efficacy of immunotherapies.
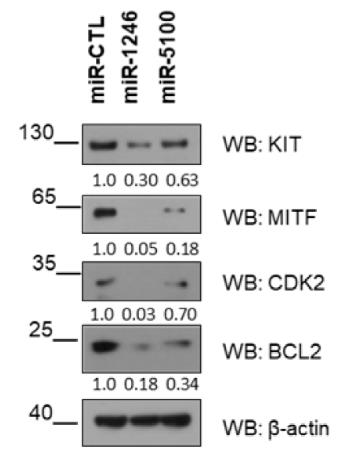
Fig3. MITF-dependent targets were assessed by western blot.
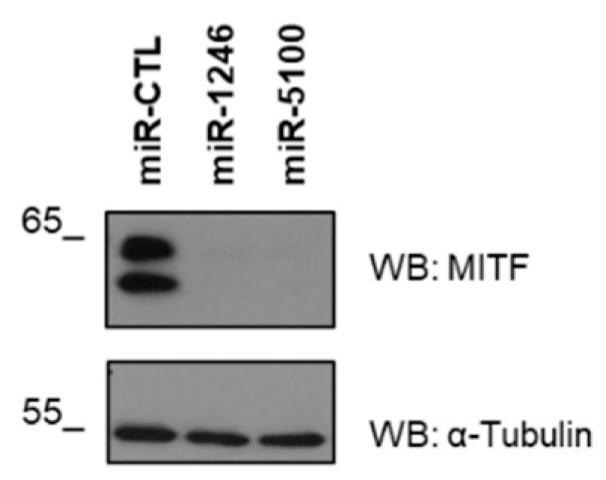
Fig4. Western blot shows MITF levels in miR-1246 and miR-5100 overexpressed LAD2.
Quality Guarantee
High Purity
.jpg)
Fig1. SDS-PAGE (MITF-5356H)
.
.jpg)
Fig2. SDS-PAGE (MITF-2446H)
Involved Pathway
MITF involved in several pathways and played different roles in them. We selected most pathways MITF participated on our site, such as Osteoclast differentiation,Melanogenesis,Pathways in cancer, which may be useful for your reference. Also, other proteins which involved in the same pathway with MITF were listed below. Creative BioMart supplied nearly all the proteins listed, you can search them on our site.
| Pathway Name | Pathway Related Protein |
|---|---|
| Melanoma | MAPK1,FGF5,PIK3CA,AKT2,FGF13,PDGFA,AKT1,BRAF,IGF1,FGF8 |
| Osteoclast differentiation | PIK3R5,PIK3CG,FHL2,MAP2K7,IL1B,MAPK12,IFNB1,FOSL2,PIK3CA,CYBB |
| Pathways in cancer | HSP90AA1,GLI1,BIRC2,FGF1,MET,ITGB1,RAD51,CTNNA3,ITGA2,PLCB3 |
| Melanogenesis | CAMK2D2,WNT2BB,WNT9A,CALM2,WNT1,WNT4A,CALM,GNA11A,FZD6,CAMK2B |
Protein Function
MITF has several biochemical functions, for example, RNA polymerase II core promoter proximal region sequence-specific DNA binding,RNA polymerase II core promoter sequence-specific DNA binding,chromatin binding. Some of the functions are cooperated with other proteins, some of the functions could acted by MITF itself. We selected most functions MITF had, and list some proteins which have the same functions with MITF. You can find most of the proteins on our site.
| Function | Related Protein |
|---|---|
| transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding | LEF1,OLIG2,HIF1A,RFX3,POU3F3,CEBPB,MYF5,RELA,TLE4,MEIS1 |
| transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding | E4F1,ASCL3,BCL6A,PURB,BCL6,CHCHD3,FEZF2,ISX,NFKB1,GLIS1 |
| protein dimerization activity | MYC,HELT,GARS,DAO1,HER2,GGN,HER8A,TFAP2C,ATM,ARNTL |
| RNA polymerase II core promoter proximal region sequence-specific DNA binding | NR3C1,SFPI1,BCL11B,CHD7,TCF7L2,MYF6,TXK,MAFB,CARF,RFX5 |
| RNA polymerase II core promoter sequence-specific DNA binding | SUZ12,SOX3,SOX11,NLRC5,YAP1,H3F3B,RBPJ,TCF7L2,SP1,MTF1 |
| chromatin binding | SIN3B,CBFA2T3,SKIL,WACA,TPR,PRKAA1,FOXN4,SP110,PYGO2,ZNF143 |
| protein binding | PPP6C,Tnfrsf22,DSP,MYRIP,PAX3,RBM5,SH3PXD2B,DDX31,GTPBP2,WRAP53 |
| transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding | BARHL2,NEUROD6,ZFAT,POU4F1,NR4A3,MAFA,ARID3B,TFAP2C,FOS,ELK1 |
Interacting Protein
MITF has direct interactions with proteins and molecules. Those interactions were detected by several methods such as yeast two hybrid, co-IP, pull-down and so on. We selected proteins and molecules interacted with MITF here. Most of them are supplied by our site. Hope this information will be useful for your research of MITF.
HK3;EBP;STUB1;YWHAZ
Resources
Research Area
Mast CellTranscription Factors in the Akt Pathway
Cancer Stem Cell Transcription Factors
Epithelial Stem Cell Transcription Factors
Melanoma Biomarkers
Oncoprotein-Transcription Factors
Related Services
Related Products
References
- Moch, H; Montironi, R; et al. Oncotargets in Different Renal Cancer Subtypes. CURRENT DRUG TARGETS 16:125-135(2015).
- Nishida, H; Suzuki, H; et al. Blockade of CD26 Signaling Inhibits Human Osteoclast Development. JOURNAL OF BONE AND MINERAL RESEARCH 29:2439-2455(2014).


