CRIP2
-
Official Full Name
cysteine-rich protein 2 -
Overview
CRIP2 and the closely related CRIP1 are cysteine-rich proteins containing two LIM domains. They are highly expressed during cardiovascular development and act to bridge serum response factor and GATA proteins and stimulate smooth muscle target genes. -
Synonyms
CRIP2;cysteine-rich protein 2;CRP2;ESP1;CRP-2;Cystein-rich intestinal protein;CRIP
Recombinant Proteins
- Zebrafish
- Rat
- Human
- Mouse
- Mammalian Cell
- Wheat Germ
- E.coli
- Insect Cell
- HEK293
- Insect Cells
- In Vitro Cell Free System
- HEK293T
- His
- GST
- Non
- His&Fc&Avi
- Myc&DDK
- His&GST
- His&SUMO
| Cat.# | Product name | Source (Host) | Species | Tag | Protein Length | Price |
|---|---|---|---|---|---|---|
| CRIP2-12660Z | Recombinant Zebrafish CRIP2 | Mammalian Cell | Zebrafish | His |
|
|
| CRIP2-1600R | Recombinant Rat CRIP2 Protein | Mammalian Cell | Rat | His |
|
|
| CRIP2-1875H | Recombinant Human CRIP2 Protein, GST-tagged | Wheat Germ | Human | GST |
|
|
| CRIP2-3051H | Recombinant Human Cysteine-rich Protein 2, His-tagged | E.coli | Human | His | 1-208 a.a. |
|
| CRIP2-491H | Recombinant Human CRIP2, None tagged | Insect Cell | Human | His | 1-208 a.a. |
|
| CRIP2-8657H | Recombinant Human CRIP2, His-GST tagged | Insect Cell | Human | His | 1-208 a.a. |
|
| CRIP2-001HCL | Recombinant Human CRIP2 cell lysate | Insect Cell | Human | Non |
|
|
| CRIP2-002HCL | Recombinant Human CRIP2 cell lysate | Insect Cell | Human | Non |
|
|
| CRIP2-1238H | Recombinant Human CRIP2 protein, GST-tagged | E.coli | Human | GST | 1-208 aa |
|
| CRIP2-1257R | Recombinant Rat CRIP2 Protein, His (Fc)-Avi-tagged | HEK293 | Rat | His&Fc&Avi |
|
|
| CRIP2-1257R-B | Recombinant Rat CRIP2 Protein Pre-coupled Magnetic Beads | HEK293 | Rat |
|
||
| CRIP2-1987H | Recombinant Human CRIP2 protein | Insect Cells | Human | Non | Met1-Pro208 |
|
| CRIP2-2056HF | Recombinant Full Length Human CRIP2 Protein, GST-tagged | In Vitro Cell Free System | Human | GST | Full L. 208 amino acids |
|
| Crip2-2312M | Recombinant Mouse Crip2 Protein, Myc/DDK-tagged | HEK293T | Mouse | Myc&DDK |
|
|
| CRIP2-2378H | Recombinant Human CRIP2 protein(Met1-Pro208), His&GST-tagged | Insect Cells | Human | His&GST | Met1-Pro208 |
|
| CRIP2-2733H | Recombinant Human CRIP2 protein, His-SUMO-tagged | E.coli | Human | His&SUMO | 1-208aa |
|
| CRIP2-2769H | Recombinant Human CRIP2 Protein, Myc/DDK-tagged, C13 and N15-labeled | HEK293T | Human | Myc&DDK |
|
|
| CRIP2-3325H | Recombinant Human CRIP2 Protein, MYC/DDK-tagged | HEK293 | Human | Myc&DDK |
|
Background
What is CRIP2 protein?
CRIP2 (cysteine rich protein 2) gene is a protein coding gene which situated on the long arm of chromosome 14 at locus 14q32. It is a putative transcription factor that contains two LIM zinc-binding domains. This protein may be involved in the differentiation of smooth muscle tissue. Additionally, CRIP2 has been identified as a nuclear copper-binding protein that regulates autophagy activation. It interacts with Atox1 in the nucleus, and copper transfer from Atox1 to CRIP2 induces a change in CRIP2's secondary structure, leading to its ubiquitin-mediated proteasomal degradation. The depletion of CRIP2 or copper-induced degradation of CRIP2 can elevate reactive oxygen species (ROS) levels and activate autophagy in certain cells. The CRIP2 protein is consisted of 208 amino acids and its molecular mass is approximately 22.5 kDa.
What is the function of CRIP2 protein?
CRIP2 is a putative transcription factor containing two LIM zinc-binding domains, which may be involved in the regulation of gene expression. The encoded protein by CRIP2 gene may participate in the differentiation of smooth muscle tissue. CRIP2 has been identified as a nuclear copper-binding protein that interacts with Atox1 in the nucleus. It plays a role in the regulation of autophagy activation, where copper transfer from Atox1 to CRIP2 induces a change in CRIP2's structure leading to its degradation, which in turn affects autophagy and reactive oxygen species (ROS) levels. CRIP2 is reported to be involved in cell fate specification and actin organization in the heart.
CRIP2 Related Signaling Pathway
CRIP2 is associated with VEGFA-VEGFR2 signaling pathways, which are important for angiogenesis and tumor growth. CRIP2 acts as a repressor of NF-kappaB-mediated proangiogenic cytokine transcription to suppress tumorigenesis. A study reported that HOXA9 inhibits HIF-1α-mediated glycolysis through interacting with CRIP2. CRIP2 has been identified as a nuclear copper-binding protein that interacts with Atox1 in the nucleus. Copper transfer from Atox1 to CRIP2 induces a change in CRIP2's secondary structure, leading to its ubiquitin-mediated proteasomal degradation.
CRIP2 Related Diseases
CRIP2 has been identified as a potential tumor suppressor gene. Its dysregulation or mutations may be linked to the development and progression of various types of cancer, including non-small cell lung cancer (NSCLC). Given its role in cardiovascular development and the differentiation of smooth muscle tissue, abnormalities in CRIP2 expression or function could potentially contribute to cardiovascular diseases. While not directly stated, proteins that regulate autophagy and inflammation, like CRIP2, could have indirect implications in autoimmune diseases where these processes are dysregulated. The role of CRIP2 in the regulation of autophagy and ROS levels suggests it may also be relevant to neurodegenerative diseases where these pathways are implicated. Since CRIP2 is involved in cell fate specification and actin organization, it could potentially be linked to developmental disorders if its function is disrupted during embryogenesis.
Bioapplications of CRIP2
CRIP2 has been associated with the radioresistance of non-small cell lung cancer (NSCLC) and could be a potential therapeutic target to improve the efficacy of radiotherapy. Due to its association with various cancers, CRIP2 could potentially be used as a diagnostic or prognostic biomarker to assess the risk or progression of the disease. Antibodies targeting CRIP2 can be used in several scientific applications, such as Western Blot, Immunocytochemistry, Immunohistochemistry, and ELISA, which are important tools for detecting and studying the protein's expression and function in various cell types and tissues.
Case Study
Case Study 1: Feifei Li, 2021
Radiotherapy plays a major role in non-small cell lung cancer (NSCLC) treatment. The curative efficacy of advanced NSCLC is unsatisfactory because of its radioresistance to conventional radiotherapy. The biomarkers which can be used to diagnose radiosensitivity or predict for prognosis are beneficial in promoting curative effects. In this study, NSCLC cell lines with acquired radioresistance to X-rays were obtained through fractionated irradiation. The differentially expressed proteins (DEPs) between the self-established radioresistant NSCLC cell line A549-R11 and control (A549-CK) were measured by proteomic analysis. Among the detected DEPs, CRIP2, ARHGDIB, and PADI3 were validated to be up-regulated in radioresistant cells, in mRNA and protein levels. Further analysis of bioinformatics deciphered that CRIP2, as a potential biomarker for diagnosis and a key biomarker for prediction of prognosis, may impact the X-ray radiosensitivity of NSCLC by regulating the occurrence of apoptosis and cell cycle arrest; as such, it may serve as a potent therapeutic target to facilitate NSCLC radiotherapy. CRIP2 and other DEPs may shed new light on the recognition of complex factors associated with radiation-responsiveness and finally be beneficial in the advancement of personalized therapies and precision medical treatment.

Fig1. Expression of CRIP2 in patients recorded in TCGA database with radioresistance or radiosensitivity.
 curve.jpg)
Fig2. CRIP2 diagnosis by receiver operating characteristic (ROC) curve.
Case Study 2: Céline Hoffmann, 2016
A critical process underlying cancer metastasis is the acquisition by tumor cells of an invasive phenotype. At the subcellular level, invasion is facilitated by actin-rich protrusions termed invadopodia, which direct extracellular matrix (ECM) degradation. Here, the researchers report the identification of a new cytoskeletal component of breast cancer cell invadopodia, namely cysteine-rich protein 2 (CRP2). They found that CRP2 was not or only weakly expressed in epithelial breast cancer cells whereas it was up-regulated in mesenchymal/invasive breast cancer cells. In addition, high expression of the CRP2 encoding gene CSRP2 was associated with significantly increased risk of metastasis in basal-like breast cancer patients. CRP2 knockdown significantly reduced the invasive potential of aggressive breast cancer cells, whereas it did not impair 2D cell migration. In keeping with this, CRP2-depleted breast cancer cells exhibited a reduced capacity to promote ECM degradation, and to secrete and express MMP-9, a matrix metalloproteinase repeatedly associated with cancer progression and metastasis. In turn, ectopic expression of CRP2 in weakly invasive cells was sufficient to stimulate cell invasion. Both GFP-fused and endogenous CRP2 localized to the extended actin core of invadopodia, a structure primarily made of actin bundles. Purified recombinant CRP2 autonomously crosslinked actin filaments into thick bundles, suggesting that CRP2 contributes to the formation/maintenance of the actin core.
 polymerized in the presence of recombinant CRP2.jpg)
Fig3. Actin filaments (1 μM) polymerized in the presence of recombinant CRP2.
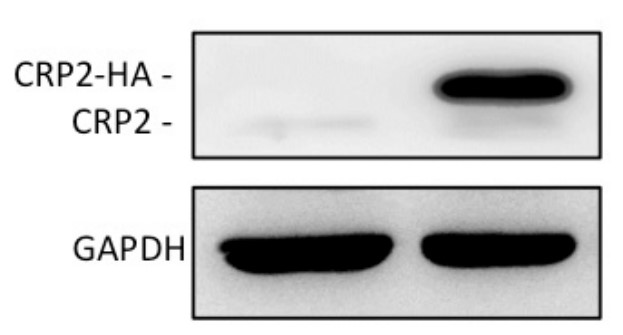
Fig4. CRP2 protein level in MCF-7 cell lines.
Quality Guarantee
High Purity
.jpg)
Fig1. SDS-PAGE (CRIP2-1875H)
.
.jpg)
Fig2. SDS-PAGE (CRIP2-2769H)
Involved Pathway
CRIP2 involved in several pathways and played different roles in them. We selected most pathways CRIP2 participated on our site, such as , which may be useful for your reference. Also, other proteins which involved in the same pathway with CRIP2 were listed below. Creative BioMart supplied nearly all the proteins listed, you can search them on our site.
| Pathway Name | Pathway Related Protein |
|---|
Protein Function
CRIP2 has several biochemical functions, for example, zinc ion binding. Some of the functions are cooperated with other proteins, some of the functions could acted by CRIP2 itself. We selected most functions CRIP2 had, and list some proteins which have the same functions with CRIP2. You can find most of the proteins on our site.
| Function | Related Protein |
|---|---|
| zinc ion binding | THRB,AKT1S1,MARCH1,ZNF212,TRIM2A,APIP,TRIM71,CPXM2,ASTL,NR6A1 |
Interacting Protein
CRIP2 has direct interactions with proteins and molecules. Those interactions were detected by several methods such as yeast two hybrid, co-IP, pull-down and so on. We selected proteins and molecules interacted with CRIP2 here. Most of them are supplied by our site. Hope this information will be useful for your research of CRIP2.
SPRY2;GADD45G;TK1;ATP1B1;ATXN1;ATN1;KLF10;SMN1;PCYT2;OSGEP;MYO1C
Resources
Related Services
Related Products
References


