H2B Family
Related Symbol Search List
- H2BFM
- H2BFWT
- HIST1H2BA
- HIST1H2BB
- HIST1H2BC
- HIST1H2BD
- HIST1H2BE
- HIST1H2BF
- HIST1H2BG
- HIST1H2BH
- HIST1H2BI
- HIST1H2BJ
- HIST1H2BK
- HIST1H2BL
- HIST1H2BM
- HIST1H2BN
- HIST1H2BO
- HIST2H2BE
Immunology Background
Background
Histone H2B is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and long N-terminal and C-terminal tails, H2B is involved with the structure of the nucleosomes. The histone H2B family proteins plays a crucial role in packaging DNA into a compact and organized structure called chromatin.
The histone H2B family is made up of 214 members from many different species, including humans. In humans, 23 genes code for histone H2B, which are located in histone clusters on chromosomes 1 and 6. These genes share a promoter region with sequences that code for histone H2A.
Histone H2B is a protein that's about 14 kDa and is made up of around 126 amino acid residues, though variants may differ. Histone H2B, along with other core histones, form dimers that come together to form an octameric nucleosome core. The nucleosome is the smallest unit of chromatin structure, which is made up of 147 bp of DNA wrapped around the core histone octamer. Histones act as spools around which DNA winds, preventing it from becoming tangled and protecting it from damage. They also play important roles in gene regulation and DNA replication.
Histone variants are usually encoded by single genes that have both introns and polyA tails. They can differ from each other in sequence, length, and expression patterns. Histone variants can also confer different structural properties on nucleosomes and have distinct functions in cell division, transcription, DNA repair, differentiation, and chromatin remodeling.
Structure
Histone H2B proteins consist of a structured histone fold domain, which is highly conserved among different species. The histone fold domain consists of three alpha-helices (α1-α3) and two short loops (L1-L2). This domain forms the core of the nucleosome and facilitates the interaction between adjacent histones and DNA. Histone H2B proteins also have variable N-terminal and C-terminal tails that are subject to various post-translational modifications, such as acetylation, methylation, phosphorylation, and ubiquitination.
Functions
| Functions | Details |
|---|---|
| Chromatin Structure | Histone H2B, along with other histones, helps in organizing DNA by forming nucleosomes. Nucleosomes consist of DNA wrapped around a histone octamer, which includes two copies each of histones H2A, H2B, H3, and H4. Histone H2B participates in stabilizing the nucleosome structure and regulating the access of proteins involved in gene expression and DNA replication to the underlying DNA. |
| Gene Expression Regulation | Modifications of histone H2B, such as acetylation, methylation, and ubiquitination, play a role in controlling gene expression. These modifications can affect the accessibility of DNA to transcription factors and the transcriptional machinery, either by promoting or inhibiting gene expression. |
| Chromatin Remodeling | Histone H2B variants, such as H2B.Z, are involved in chromatin remodeling processes. H2B variants can replace canonical H2B in nucleosomes, leading to changes in chromatin structure and gene expression patterns. |
| DNA Repair | Histone H2B participates in DNA repair processes by facilitating the recruitment of repair factors to damaged DNA sites and promoting efficient repair of DNA lesions. |
| Epigenetic Inheritance | Histone modifications on H2B, along with other histones, can be passed on during cell division, contributing to epigenetic inheritance. These modifications can influence gene expression patterns in daughter cells, even without changes in the underlying DNA sequence. |
In summary, the histone H2B family plays a critical role in chromatin structure, gene expression regulation, chromatin remodeling, DNA repair, and epigenetic inheritance. The combination of its structural features and post-translational modifications contributes to the dynamic and intricate control of gene expression and genome stability in eukaryotic cells.
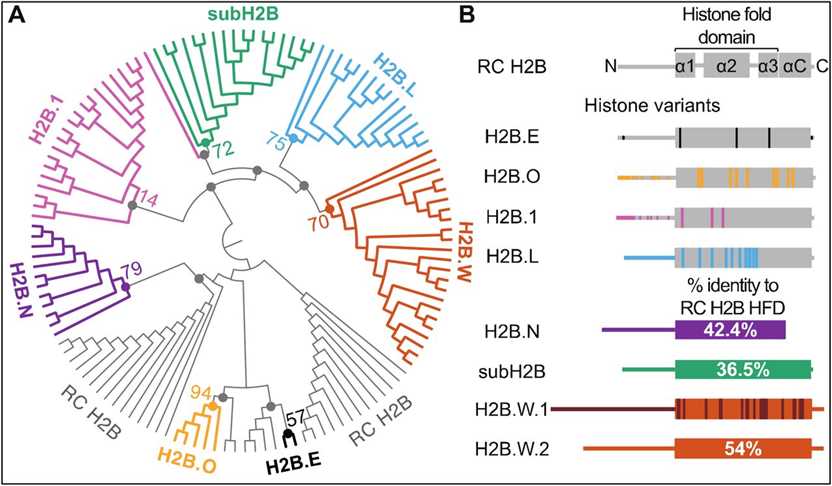 Fig.1 Phylogenomic analyses identify distinct H2B variant clades in mammals. (Pravrutha Raman, et al., 2022)
Fig.1 Phylogenomic analyses identify distinct H2B variant clades in mammals. (Pravrutha Raman, et al., 2022)Members and Variants of the H2B Family
Members of the histone H2B family include several variants that have unique functions in regulating chromatin structure and gene expression. Some of the well-known variants of histone H2B include:
| Members and variants | Details |
|---|---|
| H2B.1 | This is the canonical form of histone H2B and is found in the nucleosomes that make up chromatin. It is involved in the compact packaging of DNA and the regulation of gene expression |
| H2B.2 | This variant of histone H2B has been associated with specific cellular processes and gene regulation mechanisms. It may have distinct roles in chromatin organization and transcriptional regulation. |
| H2B.Z | Also known as H2A.Z, this variant of histone H2B is involved in the regulation of gene expression and is often associated with regions of the genome that are actively transcribed. |
| H2B.Bbd | This variant is known for its unique structural properties and its role in regulating chromatin structure and accessibility. It has been linked to specific gene expression patterns and cellular processes. |
| H2B.W | This variant of histone H2B is found in certain cell types and is thought to play a role in cell differentiation and development. |
These are just a few examples of H2B family members, and there are other variants or isoforms, such as HIST1H2BA, HIST1H2BB, HIST1H2BC, HIST1H2BD, HIST1H2BE, HIST1H2BF, HIST1H2BG, HIST1H2BH, HIST1H2BI, HIST1H2BJ, HIST1H2BK, HIST1H2BL, HIST1H2BM, HIST1H2BN, HIST1H2BO, HIST2H2BE. These variants are encoded by different genes and together constitute the dynamic structure of chromatin and play important roles in gene regulation, DNA packaging and genome stability. The specific functions and mechanisms of each member of the H2B family are currently being further investigated to elucidate their contributions to cellular processes and human health.
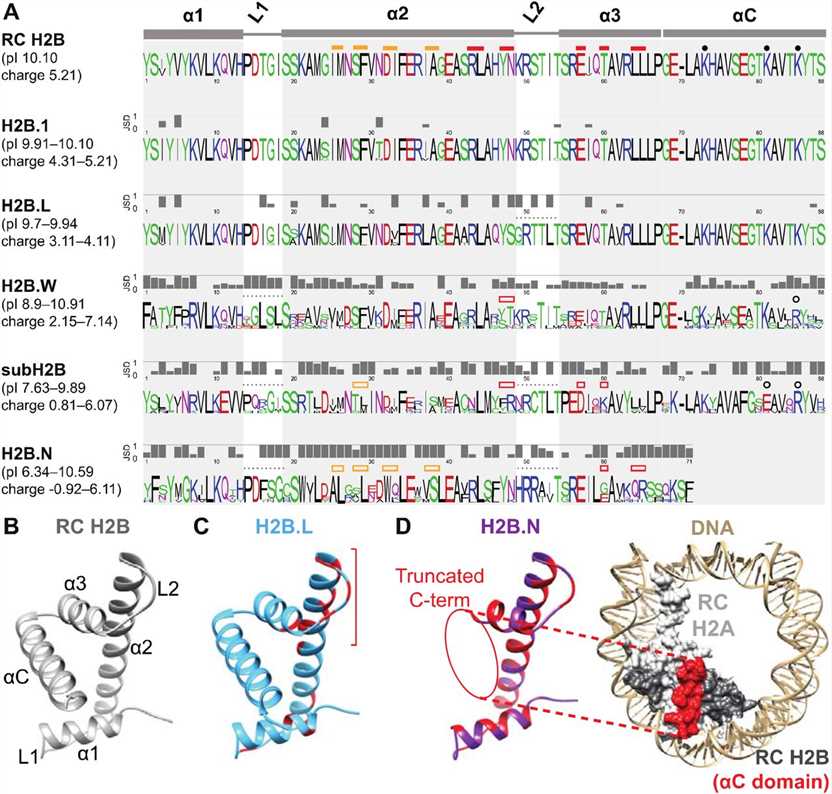 Fig.2 H2B variants diverge from RC H2B and each other in many protein features. (Pravrutha Raman, et al., 2022)
Fig.2 H2B variants diverge from RC H2B and each other in many protein features. (Pravrutha Raman, et al., 2022)Role of Histone H2B Family in Various Diseases
The histone H2B family has been implicated in various diseases, and research has made significant progress in understanding their role in disease occurrence and development. Here is an overview of the involvement of the histone H2B family in diseases and the related research progress:
Cancer
Aberrant histone modifications and alterations in chromatin structure are common features of cancer. Dysregulation of the histone H2B family members and their variants has been observed in various types of cancer. For example, altered expression of H2B variants, such as H2BFWT and H2B.Z, has been linked to tumor progression and metastasis. Research is focused on understanding the impact of these alterations on gene expression patterns, DNA repair mechanisms, and epigenetic regulation in cancer cells.
Neurological Disorders
Histone modifications and chromatin remodeling play critical roles in the development and function of the nervous system. Dysregulation of the histone H2B family has been associated with neurological disorders such as Alzheimer's disease, Parkinson's disease, and Huntington's disease. Studies are investigating the specific mechanisms by which H2B family members contribute to neurodegeneration, synaptic plasticity, and neuronal gene expression in these disorders.
Cardiovascular Diseases
Epigenetic modifications, including histone modifications, have emerged as important regulators of cardiovascular disease development. Alterations in the histone H2B family have been implicated in cardiovascular disorders such as atherosclerosis, cardiac hypertrophy, and heart failure. Researchers are exploring the impact of H2B modifications on gene expression patterns in cardiac cells and their involvement in disease progression.
Epigenetic Therapies
The dysregulation of the histone H2B family in diseases has prompted the development of epigenetic therapies. Targeting histone modifications and chromatin structure has shown promise in cancer treatment and other diseases. Histone deacetylase inhibitors (HDACis) and histone methyltransferase inhibitors (HMTis) are examples of epigenetic drugs that modulate H2B family-mediated gene expression. Ongoing research aims to optimize these therapies and identify novel targets within the H2B family for therapeutic interventions.
In summary, the histone H2B family is closely involved in the occurrence and development of various diseases. Research progress has revealed their roles in cancer, neurological disorders, cardiovascular diseases, and as targets for epigenetic therapies. Further investigations into the mechanisms underlying these associations will provide insights into disease pathogenesis and potentially lead to the development of novel therapeutic strategies.
Case Study
Case 1: Buttress T, He S, Wang L, et al. Histone H2B.8 compacts flowering plant sperm through chromatin phase separation. Nature. 2022;611(7936):614-622.
The researchers conducted a study using Arabidopsis to examine the characteristics of sperm chromatin. They utilized super-resolution 3D structured illumination microscopy (3D-SIM) to observe distinct chromatin aggregates in the sperm cells. Immunostaining for H3K9me2 revealed that larger aggregates at the sperm nuclear periphery were associated with silenced heterochromatin. These heterochromatin foci appeared enlarged compared to leaf cells. The researchers also identified a variant of histone H2B, H2B.8, which was present in sperm cells but absent in vegetative and leaf cells. Further experiments confirmed the incorporation of H2B.8 into sperm chromatin during the second pollen mitotic division, with rapid loss after fertilization and reappearance during seed maturation. H2B.8 expression was specific to sperm and seeds, consistent with previous studies. These findings suggest a unique mechanism of chromatin compaction in sperm cells and highlight the role of H2B.8 in this process.
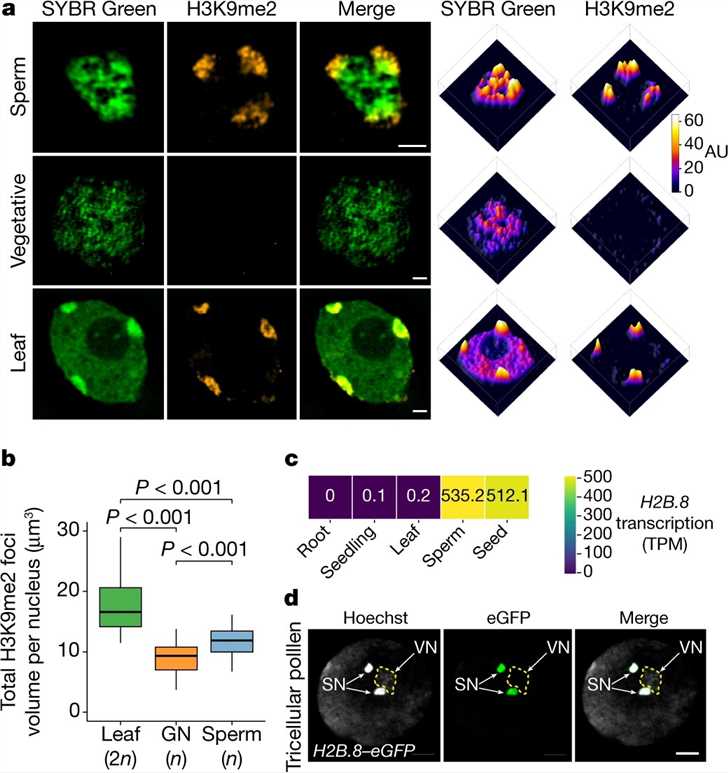 Fig.1 Sperm chromatin is aggregated and contains a specific histone variant: H2B.8.
Fig.1 Sperm chromatin is aggregated and contains a specific histone variant: H2B.8.Case 2: Nayak SR, Harrington E, Boone D, et al. A role for histone H2B variants in endocrine-resistant breast cancer. Horm Cancer. 2015;6(5-6):214-224.
In their study, the researchers aimed to investigate the role of HIST1H2BE overexpression in breast cancer cells. They utilized lentiviral infection of shRNA plasmids to lower HIST1H2BE levels in LTED cells, resulting in stable clones with decreased mRNA levels. Interestingly, total protein levels of HIST1H2B remained unchanged. The researchers observed a significant decrease in the growth rate of LTED cells, suggesting that HIST1H2BE is necessary for their full proliferation.
To determine if this effect is specific to HIST1H2BE, the researchers also decreased the expression of HIST1H2BF, another H2B variant that showed higher expression in LTED cells. However, reducing HIST1H2BF levels did not have a noticeable impact on cell growth.
Furthermore, the researchers generated stable clones of MCF-7 cells overexpressing HIST1H2BE. These clones exhibited a substantial increase in RNA expression and confirmed protein overexpression through immunoblotting. Interestingly, the HIST1H2BE overexpressing cells displayed a decreased response to estrogen stimulation. Notably, there were no alterations in estrogen receptor protein expression. Additionally, one of the overexpressing clones demonstrated increased anchorage-independent growth, while the other clone did not exhibit this effect.
In summary, the findings from this study suggest that HIST1H2BE levels play a critical role in cell growth and estrogen response in breast cancer cells. Further investigations are warranted to elucidate the precise mechanisms underlying its function, including its genome-wide incorporation into chromatin, with a particular emphasis on estrogen-regulated genes.
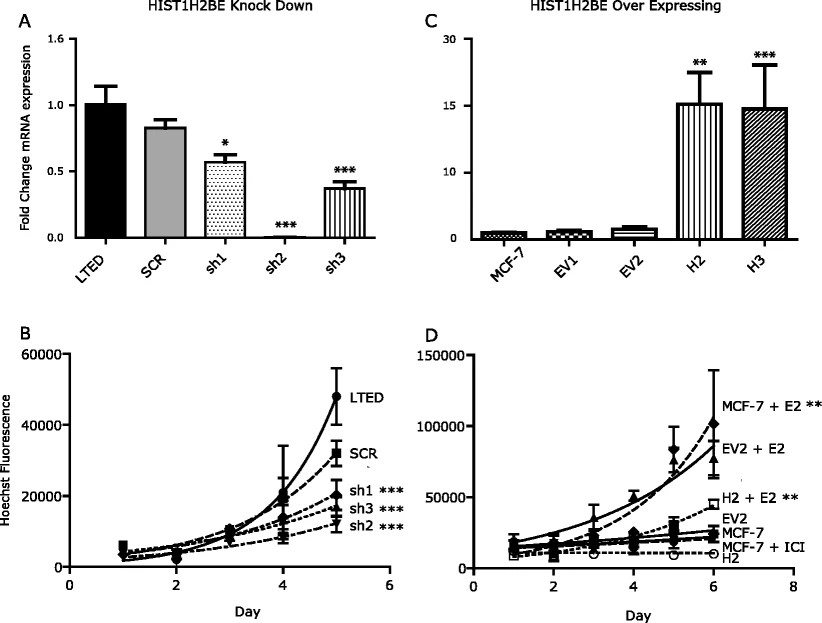 Fig.2 HIST1H2BE knockdown and overexpression affect growth of breast cancer cells.
Fig.2 HIST1H2BE knockdown and overexpression affect growth of breast cancer cells.References
- Pravrutha Raman, Mary C Rominger, Janet M Young, Antoine Molaro, Toshio Tsukiyama, Harmit S Malik, Novel classes and evolutionary turnover of histone H2B variants in the mammalian germline, Molecular Biology and Evolution, Volume 39, Issue 2, February 2022, msac019,
- Jia J, Han Z, Wang X, Zheng X, Wang S, Cui Y. H2B gene family: A prognostic biomarker and correlates with immune infiltration in glioma. Front Oncol. 2022;12:966817.
- Jiang D, Borg M, Lorković ZJ, et al. The evolution and functional divergence of the histone H2B family in plants. PLoS Genet. 2020;16(7):e1008964.
