DsbC
-
Official Full Name
-
Overview
DsbC rearranges incorrect disulfide bonds during oxidative protein folding. -
Synonyms
DsbC;thiol: disulfide interchange protein DsbC
| Cat.# | Product name | Source (Host) | Species | Tag | Protein Length | Price |
|---|---|---|---|---|---|---|
| DsbC-526E | Recombinant E.coli Thiol: Disulfide Interchange Protein DsbC | E.coli | E.coli | Non |
|
Background
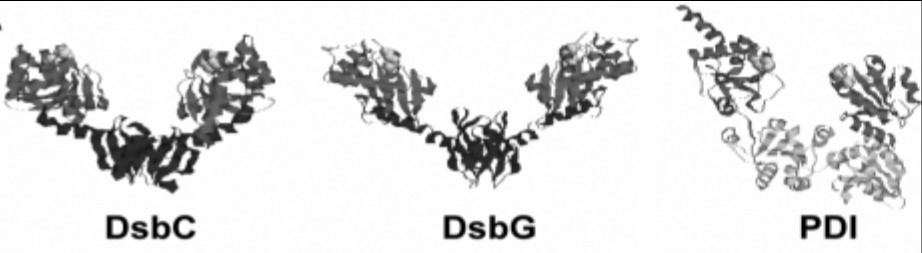
Fig1. Structures of DsbC, DsbG, and PDI exhibit similar domain arrangement. (Silvia Arredondo, 2008)
What is DSBC protein?
DsbC (thiol:disulfide interchange protein) is a protein found in bacteria that plays a key role in the proper folding of proteins and the formation of disulfide bonds. DsbC belongs to the Dsb family of proteins, members of which are responsible for maintaining the correct pairing and REDOX balance of protein disulfide bonds in bacteria such as E. coli. DsbC proteins are usually composed of two domains: an n-terminal domain and a C-terminal domain. The two domains are connected by a flexible linking region that enables DsbC to undergo conformational changes in its catalytic activity.
What is the function of DSBC protein?
DsbC is mainly located in the periplasm of bacteria, which is a region between the cell membrane and the outer wall. The main function of DsbC is to correct the faulty disulfide bonds introduced by DsbA by catalyzing the reduction and isomerization of mismatched disulfide bonds in proteins. DsbC, through its interaction with DsbA, helps maintain the correct formation of disulfide bonds in the bacterial periplasmic lumen, which is essential for the proper folding and function of secreted proteins.
DSBC Related Signaling Pathway
The role of the DsbC protein in bacteria is focused on helping the protein reach the correct folded state by forming and rearranging disulfide bonds. This process is essential for the survival and function of bacteria, especially in response to external environmental stresses such as temperature changes, oxidative stress and other conditions. In addition to its role in protein folding, DsbC may also interact with other signaling pathways, such as working in concert with other members of the Dsb family, such as DsbA, DsbB, DsbD, to coordinate the folding and quality control of bacterial proteins.
DSBC Related Diseases
Although DsbC itself is a protein in bacteria and is not associated with direct disease in humans, its role in biological research and medical applications has been associated with some diseases. The role of DsbC in protein folding suggests that it may be involved in diseases caused by abnormal protein folding. For example, neurodegenerative diseases such as Alzheimer's disease, Parkinson's disease, and Huntington's disease are all associated with the misfolding and aggregation of specific proteins.
Bioapplications of DSBC
DsbC has important applications in biotechnology and genetic engineering, especially in the expression of heteroproteins rich in disulfide bonds. Through co-expression of DsbC, the soluble expression of target protein in Escherichia coli can be increased, thus increasing the bioactivity and yield of the protein. In addition, the functional properties of DsbC make it an important tool for studying protein folding, disulfide bond formation, and REDOX equilibrium.
The role of DsbC in promoting the correct folding of proteins has important implications for vaccine design and development. For example, through DSBC-mediated protein folding, the stability and immunogenicity of pathogen antigens can be improved, which is potentially valuable for the development of vaccines against infectious diseases such as hepatitis C virus (HCV).
Case Study
Case Study 1: Jonathan L Pan, 2008
The catalytic disulfide of DsbA is regenerated by the inner-membrane protein DsbB. To help identify the specificity determinants in DsbB and to understand the nature of the kinetic barrier preventing direct oxidation of newly secreted proteins by DsbB, the researchers imposed selective pressure to find novel mutations in DsbB that would function to bypass the need for the disulfide carrier DsbA. These mutations changed hydrophobic residues into nonhydrophobic residues. The DsbB mutants were dependent on the disulfide oxidoreductase DsbC, a soluble periplasmic thiol-disulfide isomerase, for complementation. DsbB is not normally able to oxidize DsbC, possibly due to a steric clash that occurs between DsbC and the membrane adjacent to DsbB. DsbC must be in the reduced form to function as an isomerase. In contrast, DsbA must remain oxidized to function as an oxidizing thiol-disulfide oxidoreductase. The lack of interaction that normally exists between DsbB and DsbC appears to provide a means to separate the DsbA-DsbB oxidation pathway and the DsbC-DsbD isomerization pathway.

Fig1. In vivo redox state analysis of DsbC in the presence of DsbB mutants in ΔdsbABD backgrounds.

Fig2. Dimeric DsbC does not interact with DsbB due to steric clash between one protomer of DsbC with the membrane.
Case Study 2: Didier Vertommen, 2008
In Escherichia coli, DsbA introduces disulphide bonds into secreted proteins. DsbA is recycled by DsbB, which generates disulphides from quinone reduction. DsbA is not known to have any proofreading activity and can form incorrect disulphides in proteins with multiple cysteines. These incorrect disulphides are thought to be corrected by a protein disulphide isomerase, DsbC, which is kept in the reduced and active configuration by DsbD. The DsbC/DsbD isomerization pathway is considered to be isolated from the DsbA/DsbB pathway. This study shows that the DsbC and DsbA pathways are more intimately connected than previously thought. dsbA(-)dsbC(-) mutants have a number of phenotypes not exhibited by either dsbA(-), dsbC(-) or dsbA(-)dsbD(-) mutations: they exhibit an increased permeability of the outer membrane, are resistant to the lambdoid phage Phi80, and are unable to assemble the maltoporin LamB. Using differential two-dimensional liquid chromatographic tandem mass spectrometry/mass spectrometry analysis, the researchers estimated the abundance of about 130 secreted proteins in various dsb(-) strains. dsbA(-)dsbC(-) mutants exhibit unique changes at the protein level that are not exhibited by dsbA(-)dsbD(-) mutants.

Fig3. Growth curves of wild-type (□), dsbC− (▴), dsbA− (*), dsbA−dsbD− and dsbA−dsbC− (●) strains in M63 minimal media at 37°C. Growth was monitored at A600.

Fig4. In vivo redox state of DsbC. Exponentially growing cells (in LB) were TCA-precipitated, free cysteines were modified by AMS, and DsbC was detected by Western blot analysis.
Involved Pathway
DsbC involved in several pathways and played different roles in them. We selected most pathways DsbC participated on our site, such as , which may be useful for your reference. Also, other proteins which involved in the same pathway with DsbC were listed below. Creative BioMart supplied nearly all the proteins listed, you can search them on our site.
| Pathway Name | Pathway Related Protein |
|---|
Protein Function
DsbC has several biochemical functions, for example, . Some of the functions are cooperated with other proteins, some of the functions could acted by DsbC itself. We selected most functions DsbC had, and list some proteins which have the same functions with DsbC. You can find most of the proteins on our site.
| Function | Related Protein |
|---|
Interacting Protein
DsbC has direct interactions with proteins and molecules. Those interactions were detected by several methods such as yeast two hybrid, co-IP, pull-down and so on. We selected proteins and molecules interacted with DsbC here. Most of them are supplied by our site. Hope this information will be useful for your research of DsbC.
Resources
Related Services
Related Products
References


