Uncategorized Thursday, 2024/08/29
Dr. Li Wenwei from Walther Mothes’ team at Yale University, along with doctoral students Michael Grunst and Dr. Qin Zhuan, published a research paper titled ” Structure and inhibition of SARS-CoV-2 spike refolding in membranes” in the journal Science. In this study, the research team used Cryo-electron tomography (Cryo-ET) to capture the structure of COVID-19 spike protein binding to the cell receptor ACE2 in the membrane, as well as the downstream conformational changes of its bottom protein S2, revealing the antiviral mechanism of the stem helix region binding class neutralizing antibodies.
In the pre-fusion state, the spike protein trimer and ACE2 receptor dimer crosslink with each other, forming a protein array between the membranes, which helps the virus attach to the host surface. The three monomers of spike protein can each bind to one ACE2 receptor dimer, but due to structural conformational limitations, the bound dimer ACE2 protein cannot simultaneously bind to two monomers from the same spike protein trimer, but can bind to monomers from adjacent spike protein trimers, thereby achieving cross-linking of spike proteins. This cross-linking mechanism enhances the flexibility of spike-ACE2 binding to a certain extent, providing adaptive space for the virus to introduce mutation sites on spike proteins.
Our Related Proteins
After thermal activation, the spike-ACE2 complex undergoes conformational changes, forming a rod-shaped structure between the viral membrane and the host membrane. The rod-shaped structure is similar to the previously discovered post-fusion spike protein, but its direction is completely opposite – the rod-shaped structure extends from the host membrane, while the fused spike protein extends from the viral membrane. Researchers speculate that the rod-shaped structure should be the pre-hairpin intermediate state of the spike protein that they have been searching for. At this point, the bottom protein S2 has inserted the fusion peptide segment into the host membrane, forming a spiral rod-shaped structure. The bottom region of S2 exhibits high variability and cannot be stabilized by the local tomography averaging technique. Through subtomogram classification, the research team obtained spike protein intermediate states with different tilt angles on the viral membrane. These tilt angles are highly negatively correlated with the distance between the membranes, supporting the research model of spike proteins bringing the viral membrane and host membrane closer through conformational changes in self-folding, ultimately promoting double membrane fusion.
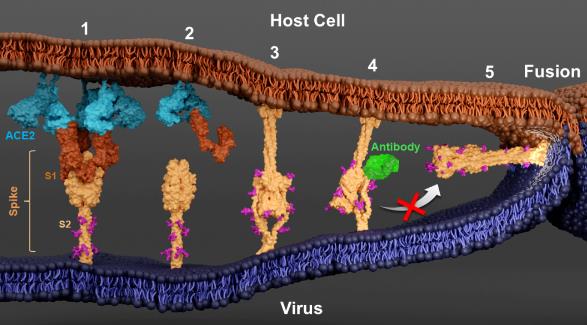
In previous studies, the research team isolated a class of neutralizing antibodies that can bind to the stem helix region at the bottom of spike proteins. Due to the high conservation of this region among coronaviruses, these antibodies have neutralizing activity against various coronaviruses, providing a theoretical basis for the design of pan-coronavirus vaccines. However, the antiviral mechanism of this type of neutralizing antibody is not yet clear. In this study, the research team confirmed that such neutralizing antibodies can bind to the stem helix region at the bottom of spike proteins, and can still bind to the intermediate state even after conformational changes occur in spike proteins. However, the distance between the virus and the host membrane cannot be further reduced, revealing that it blocks the self-folding of the base protein S2.
The teams of Paul Whitford from Northeastern University and Jose Onuchic from Rice University used molecular dynamics simulations to reproduce the process of spike protein from its pre-fusion state to binding to ACE2 receptors, activating S2 conformational changes to form an intermediate state, and finally folding itself to the fused state. After introducing neutralizing antibodies that bind to the stem helix region, the binding of the antibodies will ultimately block the self-folding of the basal protein S2. These simulation results are highly consistent with the cryo-ET structure obtained in this study.
The intermediate state structure of spike proteins exhibits highly dynamic characteristics, especially in the stem helix region. Therefore, how to design immunogens to stimulate the immune system to produce neutralizing antibodies targeting this conserved region will become an important breakthrough in the development of pan-coronavirus vaccines.
It is worth noting that Dr. Li Wenwei and Dr. Qin Zhuan from the Walther Mothes team studied the process of HIV invasion through cryo-ET last year, and also captured some structures that were not observed in previous studies. The binding of viral proteins to receptors on the in situ membrane and subsequent conformational changes are much more complex than previous research models. Cryo-ET technology provides a new possibility for in situ observation of viral membrane protein activity, filling the gap in previous structural studies that only focused on the outer membrane of proteins.
Related Products and Services
SARS-CoV-2 Proteins and Their Target Proteins
Protein Expression and Purification Services
Reference
Grunst, M. W., Z. Qin, E. Dodero-Rojas, S. Ding, J. Prévost, Y. Chen, Y. Hu, M. Pazgier, S. Wu, X. Xie, A. Finzi, J. N. Onuchic, P. C. Whitford, W. Mothes and W. Li (2024). “Structure and inhibition of SARS-CoV-2 spike refolding in membranes.” Science 385(6710): 757-765.

