Uncategorized Monday, 2025/01/20
In a new study, researchers from Westlake University in China and the California Institute of Technology in the United States designed a protein-based system in living cells that can process multiple signals and make decisions based on these signals. The relevant research results were published in the Science journal, with the title “A synthetic protein-level neural network in mammalian cells”.
They also introduced a unique term “perceptein”, which is a combination of protein and perceptron. Perceptron is a fundamental artificial neural network concept that effectively solves binary classification problems by mapping input features to output decisions.
Our Related Proteins
| Cat.No. # | Product Name | Source (Host) | Species | Tag | Protein Length | Price |
|---|---|---|---|---|---|---|
| Dendra-21 |
Purified fluorescent protein Dendra2

|
E.coli | His |
|
||
| eRFP-12 |
Purified fluorescent protein eRFP

|
E.coli | His |
|
||
| mKate-25 |
Purified fluorescent protein mKate

|
E.coli | His |
|
||
| mOrange-22 |
Purified fluorescent protein mOrange

|
E.coli | His |
|
||
| mPlum-16 |
Purified fluorescent protein mPlum

|
E.coli | His |
|
||
| mRaspberry-17 |
Purified fluorescent protein mRaspberry

|
E.coli | His |
|
||
| mStrawberry-18 |
Purified fluorescent protein mStrawberry

|
E.coli | His |
|
||
| tdTomato-19 |
Purified fluorescent protein tdTomato

|
E.coli | His |
|
||
| dsRed-4 |
Red Fluorescent Protein (dsRed)

|
E.coli | His |
|
By combining the concepts of neural network theory with protein engineering, “perceptein” represents a biological system capable of classification calculations at the protein level, similar to a basic artificial neural network. This “perceptein” circuit can classify different signals and make corresponding responses, such as determining survival or experiencing programmed cell death.
Cells naturally process various classification cues, such as stress and developmental signals, to initiate cellular functions with different outcomes. Immune cells respond to threats based on the signals they detect. The p53 signaling pathway determines whether to repair damage or self-destruct to prevent cancer.
Scientists have been working hard to build artificial systems that can replicate this decision-making process within cells. Most existing attempts rely on DNA or RNA, which may be slow and not very direct. These authors constructed their decision-making circuits using proteins, de novo synthesized protein heterodimers, and engineered proteases, rather than DNA-based systems.
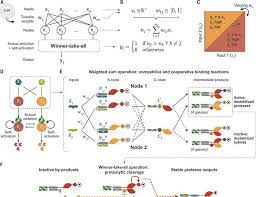
By constructing protein pairs that bind together in a specific way, these proteins arrange into a perceptein network, where some proteins activate themselves and inhibit others. This ensures that when there are multiple signals present, only the strongest signal will trigger a response, while weaker signals are ignored.
In this new study, these authors found that the perceptein circuit can distinguish signal inputs through adjustable decision boundaries, providing the possibility of controlling complex cellular responses without transcriptional regulation.
They assembled six perceptein protein components and two input proteins required to form a complete dual input dual output circuit. They chose two famous proteases: split tobacco etch virus protease and tobacco vein mottling virus protease, and fused them in a way that controlled protease cleavage and degradation.
To test the activation of the perceptein circuit, they designed a stable human embryonic kidney reporter cell line. This cell line contains a construct that simultaneously expresses two fluorescent proteins – Citrine and mCherry.
Each fluorescent protein is labeled with a cleavage-activated N-degron (degradation signal), specific to one of the two input proteases in the perceptein circuit. When the corresponding protease is in an active state, it will cleave this N-degron, reducing fluorescence. This setting allows them to intuitively and quantitatively evaluate activity based on fluorescence levels. They confirmed that each protease variant specifically reduced the fluorescence of its target reporter gene.
Further validation steps indicate that the input proteins correctly recombined their target proteases. By changing the levels of perceptein protein components, they can effectively fine-tune decision results, even with changes in input time or the introduction of noise, resulting in a strong performance.
To demonstrate practical applications, these authors connected the output of the perceptein circuit to the caspase-3 apoptotic pathway. This connection enables the perceptein circuit to trigger cell death based on specific input conditions, converting fluorescence-based outputs into cell life and death decisions.
This study demonstrates the feasibility of using synthetic proteins to construct artificial neural network-inspired circuits for complex signal classification in mammalian cells. These circuits have potential applications in programmable therapies, where cells can respond to disease-specific signals through customized outputs, such as selective apoptosis or other cellular responses.
Related Products and Services
Protein Expression and Purification Services
Reference
Zibo Chen et al. A synthetic protein-level neural network in mammalian cells. Science, 2024, doi:10.1126/science.add8468.
