Uncategorized Friday, 2024/12/20
Gene expression is the core mechanism for maintaining life activities, and translation initiation is an important link in it. The efficiency and accuracy of the translation initiation process directly affect protein synthesis, which is crucial for the normal function of cells. In eukaryotes, most mRNA molecules rely on the 7-methylguanosine cap (mRNA cap) located at the 5′ end to initiate translation. However, the specific molecular mechanism of this critical step has long been a hot and difficult topic in research.
Previous studies have shown that a trimeric complex called eIF4F plays an important role in recognizing mRNA cap ends and activating translation. EIF4F consists of three subunits: eIF4E responsible for directly binding to the cap end, eIF4A with helicase function, and eIF4G as a scaffold protein. These subunits work together to help recruit mRNA into ribosome pre-initiation complexes, thereby initiating protein synthesis. However, despite decades of research on these molecules, there are still many unanswered questions about how eIF4F efficiently and accurately identifies the cap end and how it completes the critical step of mRNA activation.
“The mechanism of mRNA cap recognition” in Nature addresses the gap in this field by developing a single-molecule fluorescence resonance energy transfer (smFRET) technique. By observing the behavior of individual molecules in real-time, the dynamic behavior of eIF4F complexes on mRNA is analyzed in detail. Research has found that eIF4F can not only randomly explore different positions of mRNA, but also exhibit unique “selectivity” and “activation” patterns in recognizing cap tips through the cooperation between its internal subunits. In addition, the study also revealed the key role of eIF4A in removing non-synthetically bound eIF4F complexes, as well as how eIF4E stabilizes the entire complex when bound to the cap end. This study not only fills the gap in translation initiation mechanisms, but also provides a new approach for developing potential disease treatment methods targeting translation regulation.
Our Related Proteins
| Cat.No. # | Product Name | Source (Host) | Species | Tag | Protein Length | Price |
|---|---|---|---|---|---|---|
| EIF4E-2775H |
Recombinant Human EIF4E, His-tagged

|
E.coli | Human | His | 1-217 aa | |
| EIF4E-2050H | Recombinant Human EIF4E protein | E.coli | Human | Non | 2-217 |
|
| EIF4E-2373H | Recombinant Human Eukaryotic Translation Initiation Factor 4E, GST-tagged | E.coli | Human | GST | 2-216 a.a. |
|
| EIF4G1-28550TH | Recombinant Human EIF4G1, GST-tagged | Wheat Germ | Human | GST | ||
| EIF4A2-12376H | Recombinant Human EIF4A2, GST-tagged | E.coli | Human | GST | C-term-300a.a. | |
| EIF4G1-12383H | Recombinant Human EIF4G1 protein, His-tagged | E.coli | Human | His | 296-645 aa | |
| EIF4E-5106M | Recombinant Mouse EIF4E Protein | Mammalian Cells | Mouse | His |
|
|
| EIF4A2-3917H | Recombinant Human EIF4A2 protein, His-tagged | E.coli | Human | His | 1-408 aa | |
| EIF4G1-01H |
Active Recombinant Human EIF4G1 protein, Myc/DDK-tagged
|
HEK293 | Human | DDK&Myc |
|
The mysterious role of the mRNA cap end
DNA contains the codes for life activities, but translating these codes into functionally diverse proteins is crucial. In this process, mRNA (messenger RNA) plays an indispensable bridging role. It not only transmits the genetic information of DNA to the “factory” of protein synthesis – ribosomes, but also requires precise regulation to ensure the efficiency and accuracy of gene expression. However, one of the core players in this process, the 7-methylguanosine cap of mRNA, hides a complex and mysterious mechanism of action.
The mRNA cap is a chemical structure carried by each eukaryotic mRNA at its 5′ end, which not only provides stability for mRNA and prevents it from being degraded by nucleases, but also signals the recruitment of ribosomes. According to statistics, over 95% of eukaryotic mRNA relies on the translation initiation process involving the cap end to achieve protein synthesis. The importance of the cap end lies not only in its physical structure, but also in the precise molecular mechanisms it participates in, which directly affects the regulation of gene expression within cells. Research has shown that the cap end functions like a “pass inspector” in the initiation of translation, determining whether an mRNA can smoothly enter the ribosome for translation.
Although the basic function of the cap has long been known, its specific mechanism of action has not been fully revealed. For example, the operation of the eukaryotic translation initiation factor 4F complex (eIF4F), which is responsible for identifying the key molecule at the cap end, has long been considered a multi-step complex process. However, how is this recognition achieved? How can molecules collaborate to complete this precise ‘hat end search’? These issues remain unresolved.
Therefore, the study of mRNA cap ends has become an important field in molecular biology. Unlocking the mystery of this mechanism not only helps us gain a deeper understanding of the core processes of gene expression, but may also provide new targets for the treatment and translation regulation of diseases such as cancer and viral infections.
Translation Gatekeeper: Complex Collaboration of eIF4F Complex
eIF4F, this molecular machine composed of three subunits works together to ensure the successful loading of mRNA into the ribosome, thereby initiating the protein translation process.
The three subunits of eIF4F complex are eIF4E, eIF4A, and eIF4G. They each perform their duties and work closely together. EIF4E is a small cap end binding protein that can specifically recognize and bind to the 7-methylguanosine cap end of mRNA, serving as the “starting point” of the entire complex. EIF4G is the core scaffold protein of the complex, which not only connects eIF4E and eIF4A, but may also interact with other translation factors to link mRNA with the 43S pre-initiation complex of ribosomes. EIF4A is a DEAD box RNA helicase that provides energy by hydrolyzing ATP, unraveling the RNA secondary structure near the cap end, and allowing ribosomes to enter more smoothly.
However, the collaboration of these three subunits is not a simple mechanical superposition, but is achieved through precise dynamic regulation. Research has found that the binding of eIF4E to the cap end can be further stabilized by eIF4G, while guiding eIF4A to carry out unwinding activities in the correct region. And this collaboration not only improves translation efficiency, but also endows the translation initiation process with a high degree of specificity. Although the working model of eIF4F has been widely accepted by researchers, there are still many unsolved mysteries regarding the specific functions and molecular mechanisms of each subunit in the translation initiation process.
Breaking through technological limitations: application of single-molecule fluorescence technology
Studying the mechanism of translation initiation at the molecular level is a complex challenge. Traditional biochemical experimental methods often rely on the average performance of large-scale samples, which makes it difficult to reveal the dynamic interactions between molecules. The limitations of this technique are particularly prominent when exploring the mRNA cap recognition mechanism and the behavior of translation initiation factor 4F complex (eIF4F). To address this challenge, researchers have introduced single-molecule fluorescence resonance energy transfer (smFRET) technology, which visualizes the fine dynamics at the molecular level.
SmFRET is a fluorescence signal-based technique that allows researchers to observe in real-time the movement and interactions of individual molecules in their environment by labeling specific sites on the molecule. In this study, the researchers placed fluorescent markers at specific locations on mRNA molecules (near or away from the cap), as well as on the key subunit eIF4G of the eIF4F complex. Through this method, they were able to accurately measure the binding and dissociation dynamics of eIF4F complexes on mRNA, even down to the binding rate and duration at different locations.
Compared to traditional polymerization experiments, the advantage of smFRET lies in its “single-molecule resolution”. The dynamic behavior of the eIF4F complex was revealed through observation of over 60000 molecules and testing under over 80 experimental conditions.
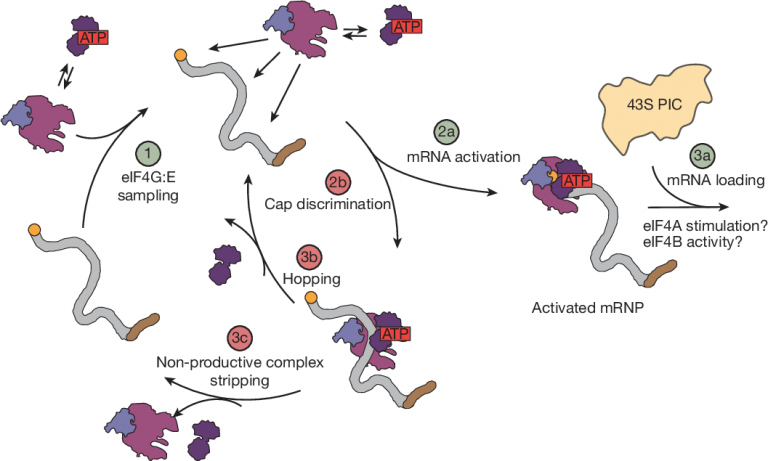
Through smFRET technology, researchers have revealed for the first time the dynamic behavior of eIF4F on mRNA, demonstrating the mysterious process from “random search” to “precise recognition”.
The study found that the recognition of the eIF4F complex is not initially locked to the cap end, but is achieved through the random binding of eIF4G to mRNA. The smFRET experiment showed that the binding and dissociation of eIF4G at different positions are random, and its binding rate and duration are not affected by the presence of the cap end. This indicates that in the initial stage, the eIF4F complex did not exhibit specificity towards the cap end. This “random search” strategy allows eIF4F to quickly scan the entire mRNA without wasting time staying at specific locations.
When the cap is present, the situation changes. EIF4E triggers a series of synergistic mechanisms through its cap end binding ability, allowing eIF4F complexes to transition from an unstable binding state to a stable binding state. Research has shown that this stabilization process relies on the collaboration between eIF4G and eIF4E: eIF4E first recognizes the cap end, and then further enhances the cap end binding ability of the entire complex through interaction with eIF4G. In addition, cap end recognition also suppresses the “scavenger” function of eIF4A to ensure that the complex already bound to the cap end is not removed.
Surprisingly, even near the cap end, eIF4F still requires some dynamic adjustment. Research has shown that the recognition of the cap end is not solely determined by chemical binding, but also involves conformational changes between subunits within the complex. This dynamic mechanism indicates that the eIF4F complex is not only a simple molecular ‘lock’, but also a dynamically responsive ‘regulatory center’.
Through these findings, researchers have proposed a new model for the cap end recognition process: eIF4F complex ensures efficiency through random exploration, triggers stability through cap end binding, and thus completes a leap from random search to precise recognition.
EIF4A’s “cleaner” role: driving the hidden power of hat end recognition
In the process of translation initiation, eIF4A is often considered as an auxiliary role, whose main function is to act as an RNA helicase to clear secondary structures near the cap end. However, the study revealed another key function of eIF4A – the “scavenger” role, which optimizes eIF4F’s precise recognition ability at the mRNA cap end by removing non-synthetic binding complexes.
Research has shown that eIF4G can non-specifically bind to multiple locations of mRNA, including regions far from the cap end. These non-synthetic combinations, while helpful for initial random searches, may also hinder the efficiency of eIF4F complexes in accurately identifying the cap end. At this point, the “cleaner” function of eIF4A becomes apparent. Through smFRET technology, researchers have found that eIF4A, driven by ATP hydrolysis, can effectively strip non-synthetically bound eIF4G from mRNA. This process ensures that the eIF4F complex does not stay in the wrong position for a long time, creating greater dynamic space for its exploration of the cap end.
Surprisingly, this “cleaner” function has a high degree of selectivity. When the eIF4F complex finally approaches the cap end, the binding between the cap end and eIF4E undergoes a novel conformational change through eIF4G, which inhibits the clearance activity of eIF4A and protects the complex that has already bound to the cap end from further removal. This clever mechanism not only improves the efficiency of hat end recognition, but also reduces resource waste during the recognition process.
Further research has found that the activity of eIF4A depends on its direct interaction with eIF4G, rather than its simple RNA unwinding function. By regulating the binding and hydrolysis of ATP, eIF4A can dynamically jump between different mRNA regions while maintaining the stability of the complex near the cap end. This “selective” behavior pattern is like an efficient cleaner, helping the eIF4F complex accurately locate its true target.
This discovery gives new meaning to the role of eIF4A in translation initiation. It is not only an RNA helicase, but also a key driving force for optimizing cap end recognition.
Activation state at the beginning of translation: proposal of a new model
This study proposes a novel molecular model through single-molecule fluorescence technology, redefining the activation process of mRNA and opening up new perspectives for our understanding of translation regulation.
Research has found that mRNA activation is not simply cap-end recognition, but a dynamic, multi-step process. During this process, the eIF4F complex (composed of eIF4E, eIF4G, and eIF4A) played a central role. Firstly, eIF4G binds to multiple locations on mRNA through random search, including areas near and far from the cap end. Subsequently, through the “scavenger” function of eIF4A, non-synthetically bound complexes were removed, ensuring that eIF4F could continue to search for the cap end.
When the eIF4F complex reaches the cap end, the binding of eIF4E to the cap end triggers a series of novel conformational changes within the complex. This change not only further stabilizes the binding at the cap end, but also sends a “stop clearing” signal to eIF4A through eIF4G, thereby protecting the binding of eIF4F complex at the cap end. This state is referred to as the “activated state” by researchers, indicating that mRNA is ready to bind to the 43S ribosome pre-initiation complex and complete the critical step of translation initiation.
Compared with the traditional “cap end activation” model, this new model emphasizes the role of dynamic regulation. The random search of the eIF4F complex, conformational changes triggered by cap end binding, and precise regulation of eIF4A together constitute this ingenious molecular machine. This dynamism ensures the efficiency and specificity of the translation initiation process, while endowing cells with the flexibility to regulate gene expression according to environmental changes.
This model has changed our understanding of translation regulation: translation initiation is not only dependent on chemical binding between molecules, but also highly regulated by dynamic interaction and synergy.
From Basic Science to Clinical Applications
The study of mRNA cap recognition mechanism not only unravels the molecular mystery of the core biological process of translation initiation, but also brings exciting possibilities for clinical applications.
In the field of tumor research, the abnormal activity of the eIF4F complex is believed to be closely related to the rapid proliferation of cancer cells. Many tumors enhance translation efficiency by overexpressing eIF4E or altering the regulatory mechanism of eIF4G, thereby promoting the expression of cancer-related genes. Based on the molecular mechanisms revealed in this study, drug design targeting eIF4F complexes will be more precise. For example, by inhibiting the binding of eIF4E to the cap end or interfering with the “scavenger” function of eIF4A, the dependence of cancer cells on the translation initiation process can be effectively blocked, thereby inhibiting tumor growth.
Furthermore, in the context of viral infections, the potential applications of this research are also highly anticipated. Many viruses rely on the translation initiation mechanism of host cells to synthesize their proteins. For example, viruses can preferentially translate viral genomes by hijacking eIF4F complexes. By regulating the dynamic behavior of eIF4F or interfering with cap end recognition mechanisms, researchers are expected to develop novel antiviral therapies targeting virus replication. This method not only has high efficiency, but also minimizes the impact on normal translation of host cells.
More broadly, this study provides new insights into the role of translation regulation in other diseases. The abnormal activity of translation initiation factors may play an important role in the occurrence of both neurodegenerative diseases and metabolic disorders. In the future, by conducting in-depth research on this mechanism and developing corresponding intervention measures, it is expected to lay the foundation for personalized treatment plans.
Related Products and Services
Protein Expression and Purification Services
Reference
Gentry, R.C., Ide, N.A., Comunale, V.M. et al. The mechanism of mRNA cap recognition. Nature (2024). https://doi.org/10.1038/s41586-024-08304-0
