With the advent and popularity of next-generation sequencing (NGS) technology, there are now many ways to analyze DNA methylation across the entire genome or large regions of the genome. These approaches open the possibility of discovering studies that quantify DNA methylation patterns and differences in DNA methylation. In addition to these hypothesis-generating methods, there is a great need for a targeted/hypothesized testing method for DNA methylation quantification. As a supplier dedicated to epigenetic research, Creative BioMart provides our customers with highly customized and cost-effective targeted bisulfite sequencing service, which allows the DNA methylation status analysis of targeted genomic regions and is very suitable for the analysis of large numbers of samples.
What Is Targeted Bisulfite Sequencing?
Targeted bisulfite sequencing is a technique based on bisulfite sequencing for assessing site-specific DNA methylation changes. This technique can accurately and effectively target any interested subset of the genome for high-resolution cytosine methylation analysis. For the purpose of “targeting”, hybridization or region-specific PCR amplification can be employed. In targeted bisulfite sequencing based on a hybridization step, bisulfite-converted DNA can be hybridized to a pre-designed oligonucleotide capture array to enrich regions of interest for targeted bisulfite sequencing, such as CpG islands, promoters, and other important methylated regions. Targeted bisulfite sequencing based on region-specific PCR amplification is performed through the bisulfite treatment of the DNA, followed by PCR amplification of the targeted region, library construction and the sequencing of the amplicon region, facilitating the methylation analysis of the single-base resolution. Specific primers are designed for regions of interest and cytosine methylation changes are evaluated in these regions.
Features of Targeted Bisulfite Sequencing
Our Advantages
Workflow of Targeted Bisulfite Sequencing at Creative BioMart
Creative BioMart strictly controls each step in the targeted bisulfite sequencing workflow to ensure access to unsurpassed data quality. In the process of targeted bisulfite sequencing based on hybridization, we utilize both commercially designed capture arrays and custom-designed capture libraries to fully meet your research objectives.
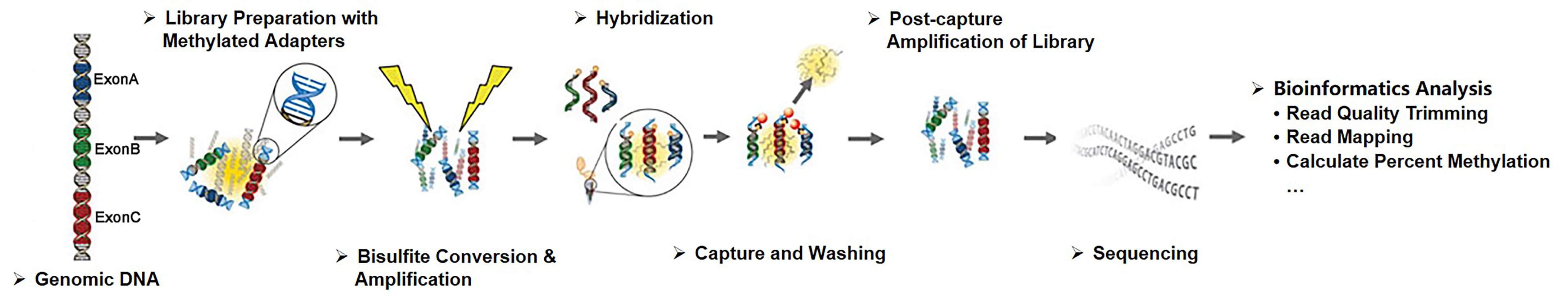
Fig 1. Workflow of targeted bisulfite sequencing based on hybridization
In the process of targeted bisulfite sequencing based on region-specific PCR amplification, the genomic DNA is bisulfite converted and multiplex amplified using specifically designed and validated primers, and the amplicons are pooled through barcoding and adapterization. The amplicon libraries are then subjected to sequencing.
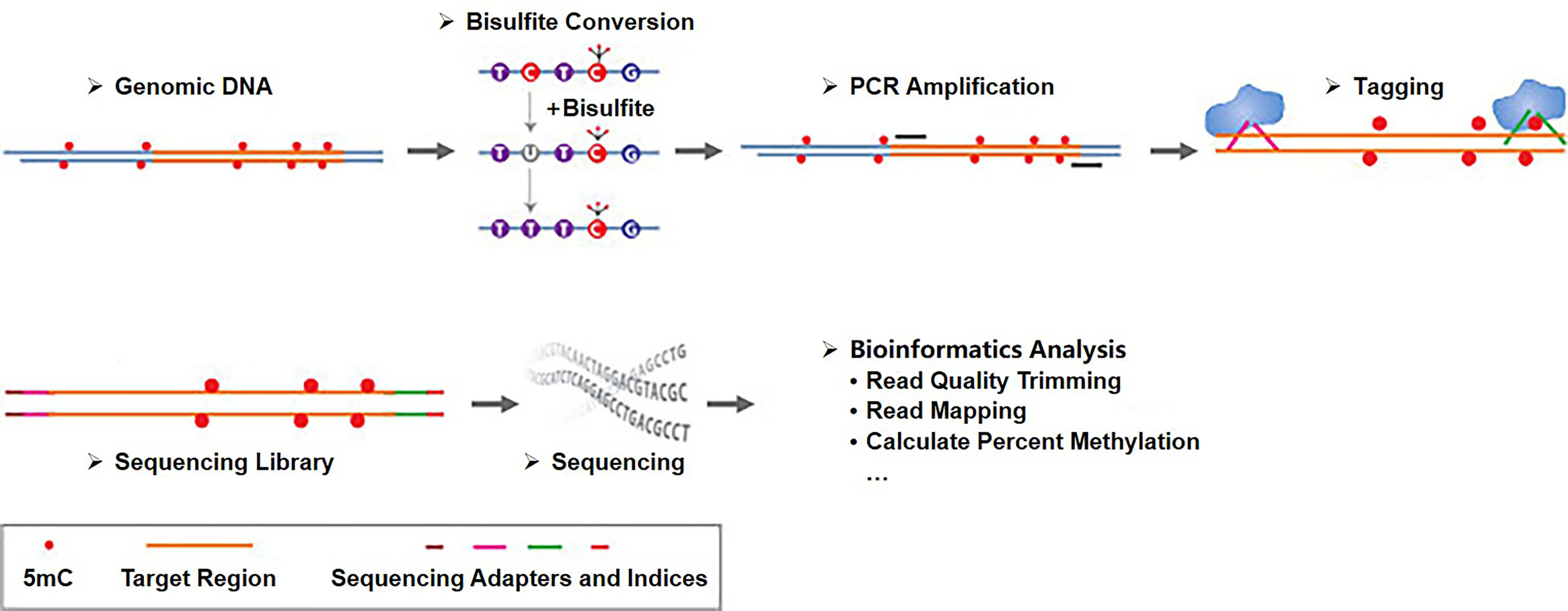
Fig 2. Workflow of targeted bisulfite sequencing based on region-specific PCR amplification
With experienced experts and first-class technical platform, Creative BioMart can provide high-quality targeted bisulfite sequencing service, ensuring the objectives are met and accelerating your epigenetic research. If you have any requirements or questions, please do not hesitate to contact us.
References
1. Li Q.; et al. Post-conversion targeted capture of modified cytosines in mammalian and plant genomes. Nucleic Acids Research. 2015, 43(12): e81.
2. Masser D R.; et al. Targeted DNA methylation analysis by next-generation sequencing. Journal of Visualized Experiments. 2015, (96): e52488.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools