In epigenetics, DNA methylation and nucleosome occupancy jointly regulate gene expression, and the analysis of these two crucial processes is of great significance to understand the genetic mechanism and the pathogenesis of diseases.
Creative BioMart currently offers NOMe-Seq (nucleosome occupancy and methylome sequencing) service, which enables simultaneous detection of DNA methylation and nucleosome/transcription factor localization within the same DNA molecule, and even provides a temporal snapshot of the relationship between nucleosome occupancy and DNA methylation.
What Is NOMe-Seq?
NOMe-Seq is the first technique to simultaneously analyze nucleosome occupancy and CpG methylation status on the same DNA strand, allowing comprehensive detect how these epigenetic factors are interrelated.
In the NOMe-Seq process, first, the fixed chromatin is treated with the GpC methyltransferase (M.CviPI) to methylate GpC dinucleotides that are not protected by nucleosomes and other proteins bound to the DNA. Due to the high abundance of GpC residues in the genome, a high-resolution footprint of nucleosome occupancy can be realized through the enzymatic treatment. The cross-links are then reversed, and the DNA is bisulfite treated. The unmethylated cytosine is converted into uracil via bisulfite conversion, followed by DNA sequencing to determine DNA methylation profiles. Because the GpC dinucleotide in the mammalian genome is unmethylated, endogenous CpG methylation can be distinguished from artificially introduced GpC methylation. By combining the sequencing information for the GpC sites with the CpG sites, nucleosome occupancy and DNA methylation of specific loci can be determined.
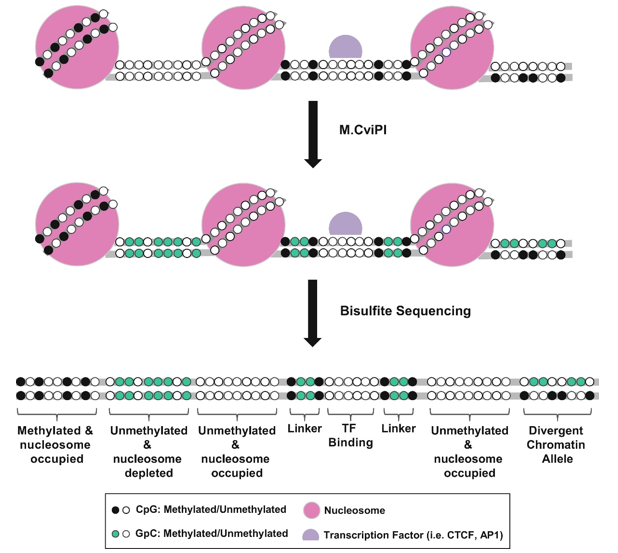
Figure 1. Schematic illustration of the principle behind NOMe-Seq. (Lay F D.; et al. 2018)
Applications & Features of NOMe-Seq
Our Advantages
Workflow of NOMe-Seq Service at Creative BioMart

Figure 2. Workflow of NOMe-Seq Service at Creative BioMart
At Creative BioMart, the enzyme-treated DNA can be analyzed in a locus-specific or genome-wide manner. If you have additional requirements or questions about our NOMe-Seq service, please don’t hesitate to contact us.
References
1. Kelly T K.; et al. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Research. 2012, 22(12): 2497-2506.
2. Lay F D.; et al. Nucleosome occupancy and methylome sequencing (NOMe-seq). DNA Methylation Protocols. 2018, 267-284.
USA
Enter your email here to subscribe.
Follow us on

Easy access to products and services you need from our library via powerful searching tools