Protein Interaction Service
Understanding how proteins interact is essential for revealing complex biological processes, like signaling pathways and finding new drug targets. At Creative BioMart, we equip researchers globally with cutting-edge techniques and expertise to pinpoint these important molecular connections accurately and efficiently.
Our Protein Interaction Services stand out with advanced platforms such as yeast two-hybrid, SPR, and FRET, offering customizable workflows tailored to your specific research objectives. Whether you’re looking to validate protein interactions, map binding sites, or carry out high-throughput screenings, we are your trusted partner in scientific innovation.
Online Inquiry
What is Protein Interaction?
Protein-Protein Interaction (PPI) refers to the process where different protein molecules form complexes through specific forces. These interactions play vital roles in cellular processes like signaling, metabolic pathways, and gene expression regulation. Delving into protein interactions helps uncover the fundamental mechanisms of life and offers potential targets for diagnosing and treating diseases.
Protein interactions are central to various cellular processes, playing pivotal roles in:
| Research Area | Description |
|---|---|
| Signal Transduction | The interactions form the backbone of pathways, helping cells respond to external cues. |
| Metabolic Regulation | Through interactions, proteins regulate metabolic pathways, ensuring cellular balance. |
| Gene Expression Control | These interactions influence transcription factors, modulating gene expression levels. |
| Disease Mechanisms | Abnormal protein interactions are linked to diseases like cancer and neurodegenerative disorders. |
What We Offer?
Our Protein Interaction Services
|
Classifications |
Methods |
Details |
|---|---|---|
|
Protein-Protein Interaction Analysis |
Yeast two-hybrid |
Genome-wide screening of protein interactions by co-expression of bait with protein libraries. |
|
Based on the split-ubiquitin protein complementation assay and detects protein interactions directly at the membrane. |
||
|
Allows rapid and convenient analysis of protein-protein interactions in transfected mammalian cells. |
||
|
In vivo technique that detects naturally formed protein complex. |
||
|
Confirm the existence of a protein-protein interaction predicted by other research techniques and identify previously unknown protein-protein interactions as an initial screening assay. |
||
| Phage display technology |
Obtain optimal protein binding by immobilization of an antigen on magnetic beads, and screening against a phage display library. |
|
|
Real-time Interaction Monitoring |
The versatile label-free BIACORE technique allows detecting and monitoring of interactions in real time. |
|
|
Quantitative and qualitative characterization of biomolecule interactions. |
||
|
Functional Studies & Characterization |
Locate the binding sites with a high degree of confidence and predict the druggability of those sites. |
|
|
Far-western blotting |
This method employs non-antibody proteins to probe the protein(s) of interest on the blot. |
|
|
Fluorescence resonance energy transfer (FRET) |
Test the direct interaction in vitro of two candidate proteins, suitable for both wild type and mutant proteins. |
|
|
Biomolecular fluorescence complementation (BiFC) |
This technique is based on the principle that two nonfluorescent fragments of a fluorescent protein dissected at appropriate site are brought together and reconstructed to fluorescence, depending on the association or interaction between the protein fused to each fragment. |
|
|
This technique is a high-throughput method used to track the interactions and activities of proteins, and determining function on a large scale. |
||
|
Genome-wide Analysis |
RNA-binding landscape mapping of interacting proteins or RNA modification sites on a genome-wide scale. |
Advanced Technologies We Use
Yeast Two-Hybrid (Y2H)
We leverage this method to screen protein-protein interactions effectively within yeast cells. It’s perfect for high-throughput screening at the genomic level, though watch out for occasional protein folding issues.
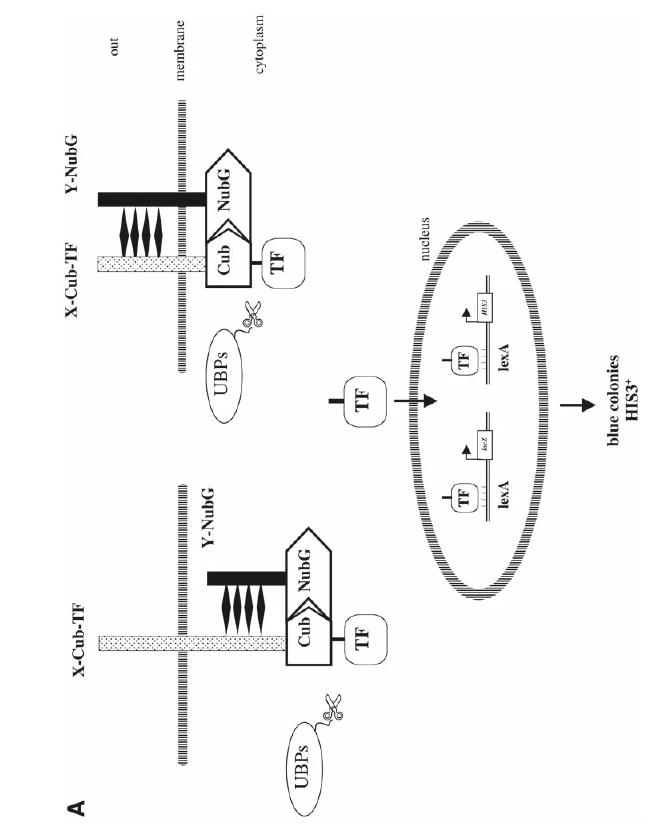
Fig. 1. The membrane-based yeast two-hybrid system. (Thaminy S.; et al. Methods Mol Biol . 2004)
This technique lets us monitor protein interactions in real-time by measuring changes in refractive index upon molecular binding. It offers precise affinity and kinetic data without the need for labeling, delivering sensitive and immediate insights.
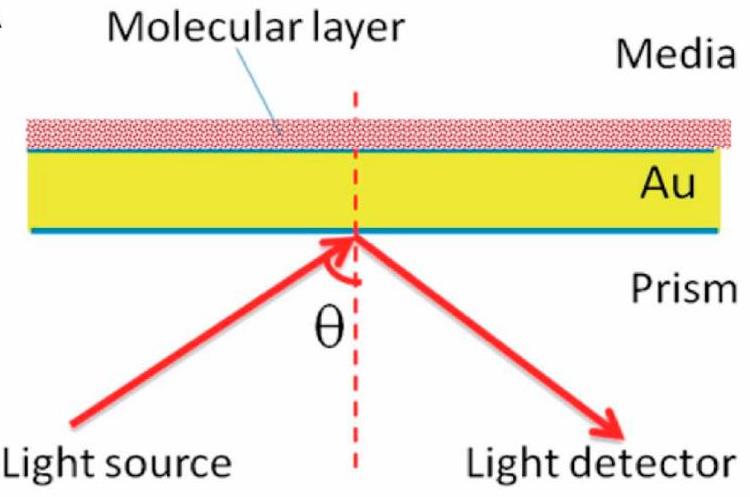
Fig. 2. Stratified medium model for surface plasmon resonance (SPR). (Li L.; et al. Biosensors (Basel). 2024)
Co-Immunoprecipitation (Co-IP)
We use Co-IP to pull down naturally occurring protein complexes from cell lysates and identify partner proteins that interact with the target. While non-specific binding can occur, we’ve honed our methods to minimize these issues.
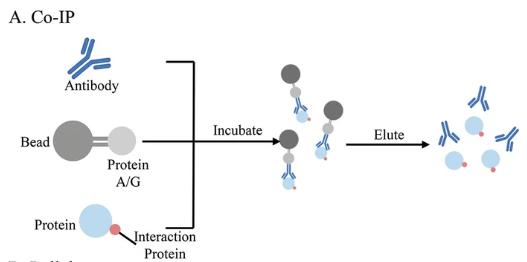
Fig. 3. Co-IP for protein-protein, protein-DNA, and protein-RNA interaction. (Yan Y.; et al. Methods Mol Biol. 2023)
X-ray Crystallography and Cryo-EM
These approaches help us dive into the 3D structures of proteins. X-ray crystallography gives atomic-level details while Cryo-EM is great for proteins that can’t be crystallized, especially large molecules and membrane proteins. Together, they provide a comprehensive view of protein interactions.
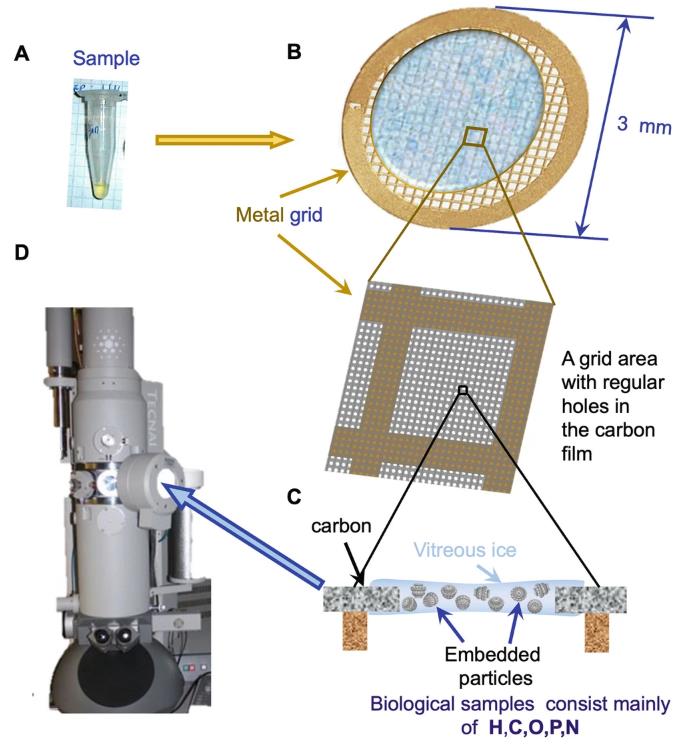
Fig. 4. Cryo-EM sample preparation. (Ignatiou A.; et al. Methods Mol Biol. 2024)
Additional Services We Offer
- Upstream Services
Target Protein Screening and Identification: Protein Engineering Services , Protein Expression and Purification Services , Membrane Proteins Expression and Purification , Protein Extraction Services , Protein Quantitation Service , Protein Characterization .
Protein Structure Analysis and Prediction: Protein Sequence Analysis and Function Prediction , Protein Network Construction and Topological Structure Analysis , Computer-aided Enzyme Design . - Downstream Services
Functional Verification and In-Depth Analysis: Gene knockout or overexpression assay, Biomarker Service , Epitope Mapping .
Drug Screening and Optimization: Drug Discovery Screening , candidate molecule optimization, and drug candidate target validation.
Applications and Benefits of Protein Interaction Services
Drug Discovery
Focus on identifying precise targets, conducting drug screenings, and exploring mechanisms.
Disease Mechanism Research
Study protein interactions to uncover the molecular basis of diseases.

Biomarker Development
Use protein interactions to identify new disease markers.
Protein Function Analysis
Reveal protein functions and predict their roles in biological processes through interaction analysis.
Service Workflow

Instrument Platform
Our company has cutting-edge tools like Surface Plasmon Resonance (SPR), mass spectrometers, Cryo-Electron Microscopy (Cryo-EM), Confocal Laser Scanning Microscope and Bio-Layer Interferometry (BLI). These instruments help us efficiently screen and analyze protein interactions, providing precise and detailed structural insights, which guarantee top-notch research results for our clients.





Case Study

Fig. 5. Two-hybrid positives from a screen of the array with a Gal4 DNA-binding domain fusion of the Pcf11 protein.
Case 1: Yeast Two-Hybrid (Uetz P.; et al. Nature . 2000)
Researchers conducted two large-scale yeast two-hybrid screens to find protein interactions in yeast, using full-length open reading frames from the Saccharomyces cerevisiae genome. First, they made an array of around 6,000 yeast samples, each with a unique activation domain fusion. This array was checked for responses in 192 proteins. In a second method, they pooled about 6,000 fusion expressions to create a library. They screened nearly all 6,000 yeast proteins, using Gal4 DNA-binding domain fusions, against this library, identifying interactions by sequence analysis. This led to 957 potential interactions among 1,004 proteins, helping place unclassified proteins in context and linking various biological functions.

Fig. 6. 3D cryo-EM map of GP38-Gn H-DS -Gc.
Case 2: Cryo-Electron Microscopy (McFadden E.; et al. Cell . 2024)
Researchers looked into CCHFV, a tickborne virus that’s quite deadly. They’ve figured out structures for GP38 and Gc glycoproteins before, but glycoprotein Gn’s structure and how it interacts were still a mystery. Using protein engineering, researchers developed a stable GP38-Gn-Gc complex and examined it with cryo-electron microscopy. This revealed how GP38 connects on the viral surface, showed Gn’s structure, and explained how GP38-Gn holds Gc’s fusion loops in place, aided by an N-linked glycan on Gn. Vaccination with this complex gave mice 40% protection against a lethal challenge. This work sets the stage for vaccine development.

Fig. 7. Generation of affinity reagents through phage display.
Case 3: Phage Display Technology (Gorman K.; et al. Curr Protoc Chem Biol. 2024)
Antibodies play a crucial role in identifying proteins in complex samples, helping to understand their location, quantity, interactions, and functions within cells. However, producing antibodies can be both time-consuming and challenging, with issues sometimes arising in their affinity or specificity. This protocol presents a quicker, cost-effective alternative by using phage display from a library of fibronectin type III (FN3) monobody variants, followed by affinity selection to create effective antibody surrogates.

Fig. 8. This analysis revealed the presence of DDB1 in the NBS1 pull-down and vice versa.
Case 4: Pull-Down Assays & Co-Immunoprecipitation (CO-IP) (Lim HE.; et al. Int J Mol Sci. 2024)
Researchers found that DNA damage-binding protein 1 (DDB1) connects with NBS1 in the DNA damage response. They used pull-down and immunoprecipitation tests in Xenopus egg extracts and human cells to show this specific interaction. It was noted that DDB1 also connects with proteins interacting with NBS1, like TopBP1 and MDC1. Importantly, without MDC1, the interaction between DDB1 and NBS1 is lost. They also observed that lacking DDB1 boosts Chk1 activation during DNA damage. These findings shed light on DDB1’s role in regulating DNA damage pathways.

Fig. 9. Loading profile comparison between IF2, GFP, PTEF-CTD, and E. coli 70S ribosome.
Case 5: Bio-layer interferometry (BLI) (Pandiarajan I.; et al. Front Mol Biosci. 2024)
The ribosome is crucial for protein translation, with its RNA core catalyzing peptide formation, aided by various proteins. Researchers study these interactions using techniques like centrifugation and light scattering. Now, they’ve added Bio-Layer Interferometry (BLI) to analyze ribosome interactions, verifying its efficiency with E. coli ribosomes and specific proteins. BLI’s advantages include label-free procedures and sample reuse, broadening its use for exploring ribosome-protein, protein-protein, and protein-ligand interactions.
Why Choose Us?
-
Advanced Equipment.
Outfitted with industry-leading tools like SPR, mass spectrometers, and Cryo-EM for efficient and accurate protein interaction analysis. -
Comprehensive Solutions.
Offer a range of experimental techniques (e.g., Y2H, Co-IP, SPR) to tailor the best research plan for clients’ needs. -
High-throughput and Precision Analysis.
Utilize automation and high-throughput tech for large-scale screening and precise analysis. -
Experienced Team.
A team of expert scientists with years of experience ensures smooth execution and optimal project outcomes. -
Customized Services.
Provide personalized experimental design and data analysis to meet diverse research goals. -
Global Service Network.
Serve clients worldwide with efficient communication and support, ensuring prompt responses and delivery.
FAQs
-
Q: How do you choose the best method for protein interaction analysis?
A: We select techniques based on research goals and needs. For instance, use yeast two-hybrid for high-throughput screening, SPR for kinetics, and Co-IP to confirm natural interactions. -
Q: What proteins can be studied in interaction research?
A: We support various protein types, like recombinant, natural, and membrane proteins, each using suitable methods for analysis and screening.
-
Q: How are protein interaction results interpreted?
A: Of course! We offer protein extraction services from small samples to high-throughput demands and customize solutions for projects of any scale.
-
Q: Is it necessary to provide protein samples for experiments?
A: Yes, clients need to supply samples of the target protein. In addition, if you do not have a ready-made protein sample, we can also provide de novo protein expression and purification services to ensure quality for interaction analysis. -
Q: What disease research benefits from protein interaction studies?
A: Protein interaction research aids in studying cancer, neurodegenerative diseases, immune disorders, infectious diseases, and more, helping identify potential targets and biomarkers. -
Q: Can drug screening and target validation be performed?
A: Yes, based on protein interaction studies, we offer services for drug screening, target validation, and new drug development.
Additional Links and Resources
Related Articles
Related Services
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions





